Towards the design of molecular materials with Dr. Brandenburg
© 2019 EPFL
On January 24 at 17:00, MED 2 1124 (CoViz2), Dr. Jan Gerit Brandenburg will present his work on crystal structure prediction, discussing both the challenge of obtaining very accurate estimates of polymorph energetics, and that of exploring the enormous search space of possible stable configurations.
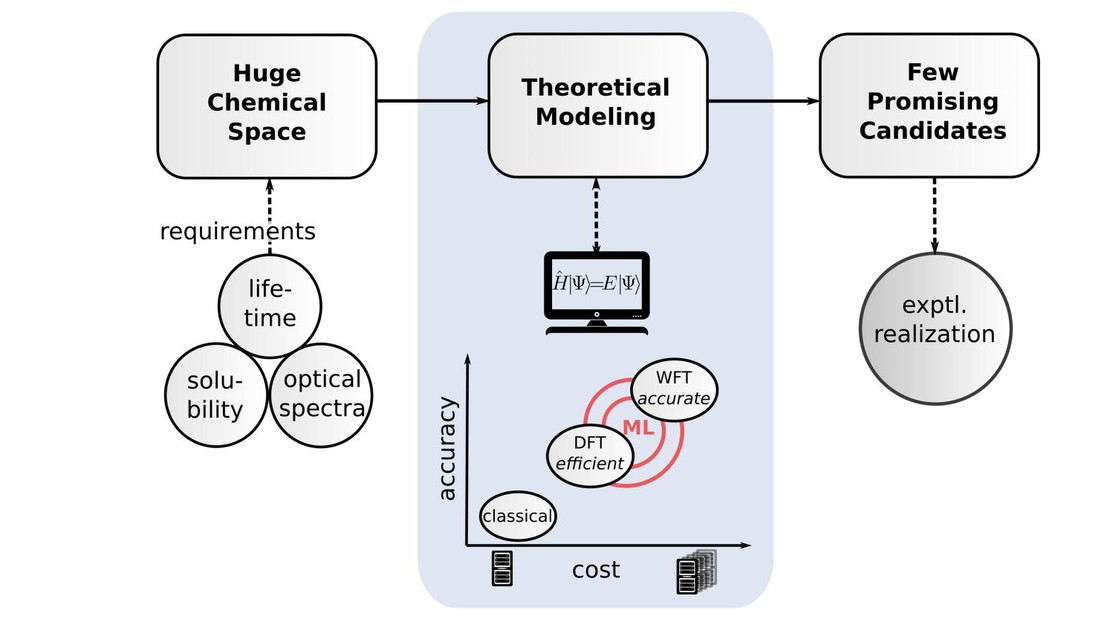
New technologies are made possible by new materials, and until recently new materials could only be discovered experimentally. However, approaches based on the fundamental laws of quantum mechanics are now integrated to many design initiatives in academia and industry (see Fig. 1), underpinning efforts such as the Materials Genome initiative or the computational crystal structure prediction (CSP [1]). The latest CSP blind test organized by the Cambridge Crystallographic Data Center [2] revealed two major remaining challenges:
(i) Crystal polymorphs are often separated by just a few kJ/mol, exceeding the accuracy of standard density functional approximations (DFAs).
(ii) Dealing with a vast search space, in particular for molecules with increased flexibility, one has to sample about 1 Mio possible crystal structures.
Recent algorithmic developments in Quantum Monte-Carlo make it feasible to molecular crystals and we are now able to predict static lattice energies with potentially sub-chemical accuracy [3]. On the other hand, cost-effective electronic structure methods will be presented that gain up to four orders of magnitude in computational speed compared to traditional DFAs and are suited for optimizing a huge number of putative crystal structures [4]. Promising applications to the CSP of pharmaceutical-like molecules have been demonstrated recently [5]. A perspective on employing machine learning techniques in the CSP context will be discussed.