Sustainable, portable and high performing – updated NEURON simulator
© 2022 EPFL
In recent years, simulation alongside theory and experimentation, has firmly become the third pillar for studying the brain. The computational models of brain components, brain tissue or even whole brains provide new ways to integrate anatomical and physiological data and allow insights into causal mechanisms crossing scales and linking structure to function.
Over the past four decades, the NEURON simulator has been developed by Michael Hines, Yale University, and continuously been extended to enable neuroscientists to model the electrical activity of neuronal networks. Today, with thousands of existing NEURON models and a large user community, it is one of the most widely used simulation environments for biologically detailed neurosimulations. As these models gain in size, complexity and detail, so do their computational requirements. In addition, these simulations are typically run on computer systems ranging from laptops to supercomputers and in the cloud, and must do so efficiently. This means that such complex software must be easily maintainable, to continuously provide these improvements crucial to the computational scientist’s work.
In a paper recently published in Frontiers in Neuroinformatics, the EPFL Blue Brain Project has collaborated with Yale University and the Department Physiology and Pharmacology, SUNY Downstate, New York to substantially modernize NEURON. The result is better software sustainability, improved software and hardware portability and increased simulator efficiency.
A focus on sustainability through modern development practices
“A key focus area has been the software sustainability of NEURON”, explains, Omar Awile, HPCEngineer at the Blue Brain Project. “NEURON’s overall code organization, testing, documentation and build system has been overhauled. Now it includes a modern build system, continuous integration and build automation and consolidated documentation generation to make it more accessible to developers and users”.
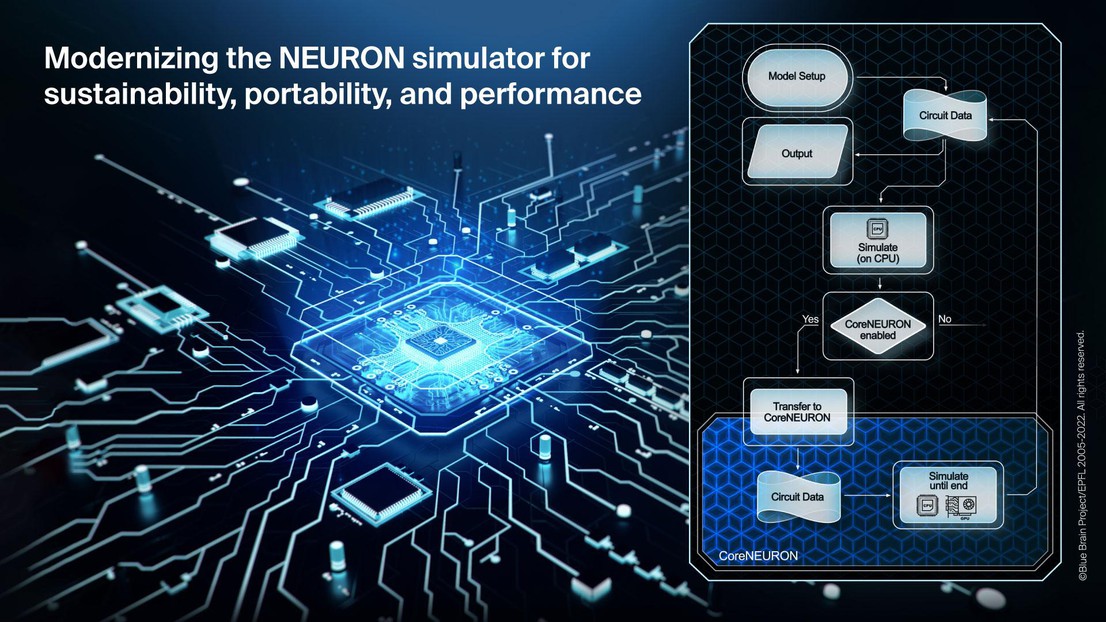
Thanks to the modern development practices introduced in this work, it also has been possible to seamlessly integrate the previously developed CoreNEURON software into NEURON along with a modern source-to-source compiler (NMODL) capable of targeting both CPUs and GPUs. CoreNEURON helps existing NEURON users to simulate their models faster, better utilizing computing resources and ultimately helps to deliver science sooner. With the achieved integration into NEURON, this is now readily available to any user. As a showcase, the paper documents the performance of several large-scale models using different multi-CPU and multi-GPU configurations.
Embracing Python’s success in scientific workflows
Python has become the dominant programming environment in scientific workflows and Python-based data processing, analysis and visualization tools are commonly used in the scientific community. Therefore, in response to its popularity, a modern NEURON Python package has been introduced. Researchers can now use NEURON’s full feature set, be it on their laptop, a cloud instance or their university’s computing cluster by simply installing its Python binary package.
“These radical updates to NEURON’s development ecosystem and our modernization efforts have already led to the adoption of a collaborative development process with a lively community,” says Prof. Felix Schürmann, Blue Brain’s Computing Director. “The growing developer base now has a simpler and more robust software distribution, a wider range of supported computer architectures, a better integration of NEURON with other scientific workflows, and substantially improved performance for the simulation of biophysical and biochemical models”.
New functionalities for NEURON
On the solid foundation of a modernized NEURON, it has also been possible to extend NEURON’s functionality and performance. In particular for sub-cellular and multiscale models, NEURON’s reaction-diffusion simulation capabilities with just-in-time compilation have been updated resulting in a more seamless integration with the rest of NEURON, support for exporting to the SBML format, and support for 3D intra- and extra-cellular simulation.
Michael Hines, Yale University, who started the development of the simulator in the 1980’s discloses what is next. “We recognize the importance of the continuity of the NEURON project to allow the neuroscience community to keep using NEURON to simulate increasingly complex brain models in more accessible ways on systems ranging from desktops to supercomputers. These updates and improvements, and welcome additions to our development team, mean we have the foundations for further modernization and re-engineering of NEURON to fulfill demand and respond to future trends”.
______________________________________
For more information, please contact Blue Brain’s Communications Manager – [email protected]
Data Availability
The NEURON simulator, with all the features and improvements described in this paper, is available as version 8.2 in the NEURON GitHub repository. https://github.com/neuronsimulator/nrn
NetPyNE with CoreNEURON support is released as version 1.0.1 and available in the NetPyNE GitHub repository. https://github.com/suny-downstate-medical-center/netpyne
The 3D Olfactory bulb model is available in the Human Brain Project GitHub repository - https://github.com/HumanBrainProject/olfactory-bulb-3d
The Rat CA1 Hippocampus model is available as part of the Hippocampus Microcircuit Massive Open Online Course12 offered on edx.org - https://www.edx.org/course/simulating-a-hippocampus-microcircuit
The M1 cortical circuit is available in the SUNY Downstate Medical Center GitHub repository - https://github.com/suny-downstate-medical-center/M1_NEURON_paper/
All the benchmarking scripts, performance measurement data and figures are available in the NEURON GitHub repository - https://github.com/neuronsimulator/neuron_frontiers_2022_artifacts21
The group behind the modernized NEURON
The collaborators involved in this study are affiliated with the following institutions:
Omar Awile, Pramod Kumbhar, Nicolas Cornu, James Gonzalo King, Olli Lupton, Ioannis Magkanaris, Fernando Pereira, Alexandru Săvulescu and Felix Schürmann - Blue Brain Project, École Polytechnique Fédérale de Lausanne, Geneva, Switzerland
Salvador Dura-Bernal, Adam J. H. Newton - Department Physiology and Pharmacology, SUNY Downstate, NY, United States
Salvador Dura-Bernal, William W. Lytton - Center for Biomedical Imaging and Neuromodulation, Nathan Kline Institute for Psychiatric Research, Orangeburg, NY, United States
Robert A. McDougal, Adam J. H. Newton - Department of Biostatistics, Yale School of Public Health, CT, United States Robert A. McDougal - Program in Computational Biology and Bioinformatics, Yale University, New Haven, CT, United States
Robert A. McDougal - Yale Center for Medical Informatics, Yale University, New Haven, CT, United States
Michael L. Hines, Nicholas T. Carnevale - Department of Neuroscience, Yale University, New Haven, CT, United States
Research reported in this publication was supported by the National Institute for Neurological Disorders and Stroke, the National Institute for Mental Health, and the National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health under award numbers R01NS011613, R01MH086638, and U24EB028998, National Science Foundation 1904444-1042C, New York State Spinal Cord Injury Research Board (SCIRB) DOH01-C32250GG- 3450000, funding to the Blue Brain Project, a research center of the Ecole Polytechnique Federale de Lausanne (EPFL), from the Swiss government’s ETH Board of the Swiss Federal Institutes of Technology, the European Union’s Horizon 2020 Framework Programme for Research and Innovation under the Specific Grant Agreement No. 785907 and 945539 (Human Brain Project SGA2 and SGA3). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health