STEPS 4.0 towards large-scale biologically-relevant simulations
© 2022 EPFL
Simulating neurons down to the level of their biochemistry in order to understand their behavior and properties is a complex computational challenge. While small to the human eye, a neuron is a comparatively large object relative to its biochemical constituents. Yet, the overall behavior of a neuron depends on the specific composition and interactions at the molecular level that will control its electrical properties. It’s like aiming to fully describe a patch of forest down to the millimeter, recording the location and shape not only of each tree and each leaf, but of each grain of dirt.
Even with high performance computing and ever increasing computing power, scientists still have to make uncomfortable compromises; phenomena that are random in nature are averaged out for ease of calculation, or only parts of a cell can be described with molecules and their interactions. Needing to choose between what is biologically relevant and what is able to be addressed, leads to significant loss of information and, often, relevance.
Now, teams from the EPFL Blue Brain Project and from the Okinawa Institute of Science and Technology Graduate University, Japan (OIST) describe how they pushed the boundaries of simulating biochemical diffusion-reaction models to the scale of entire neurons. The new solution dramatically reduces the computer memory needed while maintaining similar or better performance, increasing overall scalability.
The interdisciplinary field of computational neuroscience brings together researchers from a wide variety of backgrounds. Addressing the scientific questions posed in the field not only requires both a deep understanding of the biological system and its physical properties, but also innovative engineering solutions to hard problems in computer science and software engineering.
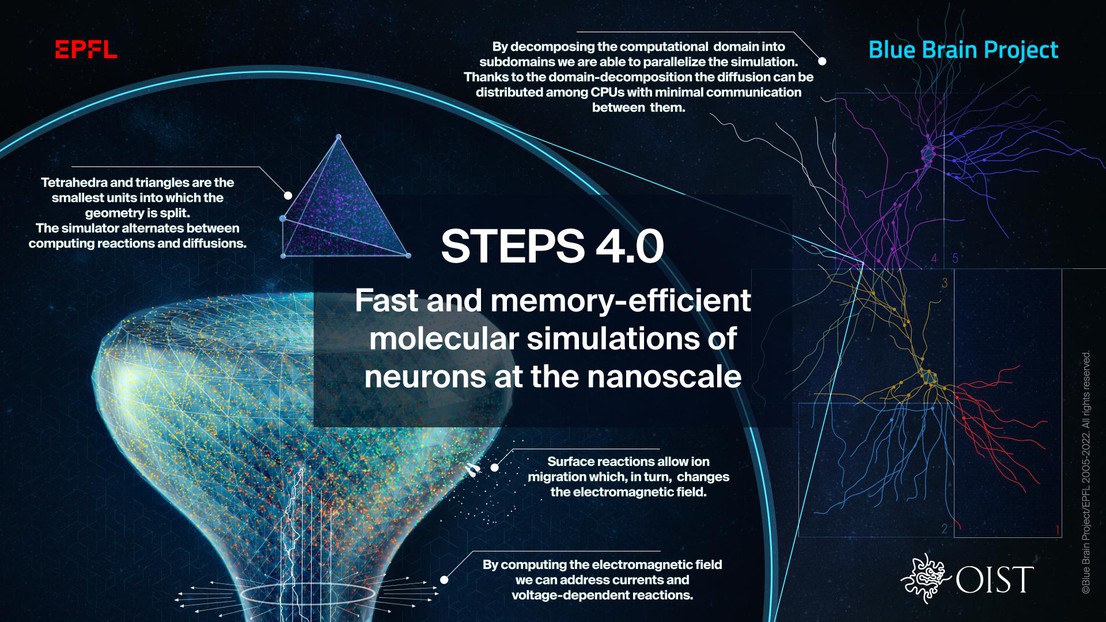
Due to complex computing difficulties, most currently used models abstract neurons as branched cables associated with ordinary differential equations describing their voltage changes. In this approach, many small structures such as axons and dendrites are hard to model, or, as in the case of spines, often ignored entirely. These simplified models have a spatial resolution in the micrometer range, which is acceptable when working with experimental data obtained from light microscopy; however, advances in experimental techniques have resulted in a deluge of data at the nanoscale. An approach that can handle complex morphologies with a resolution as low as 50 nm is therefore necessary. Additionally, the interactions between molecules are probabilistic, and any deterministic modeling would ignore stochastic effects. To further our understanding of neurons and their behavior it would be beneficial to be able to integrate this detail into our computational models.
Increasing model size and complexity require the computation of an increasingly large number of equations. However, traditional simulators running serially on laptops or workstations, as well as so-called naive parallel implementations, are inadequate to handle such detailed models.
First released publicly ten years ago, the STochastic Engine for Pathway Simulation (STEPS) has been designed to support researchers from different backgrounds and with different levels of computational expertise. It includes a friendly modeling interface that significantly reduces the need for manual coding efforts. In a paper published in Frontiers in Neuroinformatics, the Blue Brain collaboration with OIST introduces a new generation of the project - STEPS 4.0. This version of the software specifically targets large-scale modeling with complex geometries. “Although STEPS has introduced many new features and additions since its initial release in 2012, the core components and API have remained relatively unchanged,” explains Tristan Carel, HPC Engineer at the Blue Brain. “With this latest release – STEPS 4.0, the team has implemented various far-reaching changes: the core components have been redesigned for improved scalability, performance, and memory efficiency, while increasing the performance of time-critical data structures and routines. At the same time, the API has been renewed and simplified to offer a cleaner and more intuitive interface to the scientist” he confirms.
Key development towards future neuroscience research
The fundamental contribution of these efforts is the introduction of a new distributed stochastic reaction-diffusion solver supported by a sophisticated distributed mesh library. The new distributed library of volumetric computed units replaces the mesh architecture inherited from the serial approach. Since every computing core now loads only part of the mesh, the new solver greatly reduces the memory footprint of simulations while improving performance and parallel scalability, thereby resolving what was the major bottleneck in previous versions. “This improvement is essential to future neuroscience research as it enables large-scale molecular reaction-diffusion and electrophysiology simulations with biologically realistic models” explains OIST’s Weiliang Chen, Lead Author of the study.
The team evaluated the performance improvements and memory footprint of the implementation using three published models of gradually increasing complexity to cover a wide range of use cases. Starting with a simple deterministic voltage model, continuing by enriching that model with spatial stochastic reaction-diffusion and finishing with the complex, realistic test case of the calcium burst activity of a Purkinje neuron. The results from these benchmarks suggest that the latest version dramatically reduces the per-core memory consumption by more than a factor of 30, while maintaining similar or better performance and scalability.
“When we started the STEPS project ten years ago,” discloses Prof. Erik De Schutter, OIST , “it provided neuroscientists with a new tool to ask previously unanswered questions. We will continue on this track by extending STEPS 4.0 with vesicle modeling to allow even more detail. Vesicles are relatively large structures with complex molecular interactions, they play many important roles in synapses”.
Bigger, more detailed simulations that can be run for longer
Prof. Felix Schürmann, Blue Brain Computing Director, explains how Blue Brain’s research will benefit from this new capability: “With the novel features of this new enhanced version of STEPS, I am excited to see what new questions we can ask, and how we can investigate new problems through computer simulation to advance our understanding of the brain and other biological systems”. The BBP will therefore endeavor to couple STEPS with other simulator software to support multi-scale modeling and use this approach to capture elements at various temporal and spatial scales which will support the creation of large-scale reconstructions of brain tissue.
“For the Blue Brain Project, being involved in the STEPS project contributes to achieving our goals while also contributing to the community and helping to accelerate progress,” he concludes.
Data Availability Statement
The STEPS simulator is available at http://steps.sourceforge.net/. Models for validation and performance investigation presented in this publication are available at https://github.com/CNS-OIST/STEPS4ModelRelease/.
About the Authors
The collaborators involved in this study are affiliated with the following institutions:
The STEPS project is operated by Okinawa Institute of Science and Technology Graduate University (OIST), Japan: Weiliang Chen, Iain Hepburn, Jules Lallouette, Erik De Schutter
Blue Brain Project, École Polytechnique Fédérale de Lausanne (EPFL), Switzerland: Tristan Carel, Omar Awile, Nicola Cantarutti, Giacomo Castiglioni, Alessandro Cattabiani, Baudouin Del Marmol, James G King, Christos Kotsalos, Pramod Kumbhar, Samuel Melchior, Felix Schürmann
Research reported in this publication was supported by the Okinawa Institute of Science and Technology Graduate University (OIST) and funding to the Blue Brain Project, a research center of the École polytechnique fédérale de Lausanne (EPFL), from the Swiss government’s ETH Board of the Swiss Federal Institutes of Technology and the European Union’s Horizon 2020 Framework Programme for Research and Innovation under the Specific Grant Agreement No. 785907 (Human Brain Project SGA2).
Chen, W., Carel, T., Awile, O., Cantarutti, N., Castiglioni, G., Cattabiani, A., Marmol, B. D., Hepburn, I., King, J. G., Kotsalos, C., Kumbhar, P., Lallouette, J., Melchior, S., Schürmann, F., & Schutter, E. D. (2022). STEPS 4.0: Fast and memory-efficient molecular simulations of neurons at the nanoscale.https://doi.org/10.3389/fninf.2022.883742