Recognizing Local and Global Structural Motifs At the Atomic Scale
© 2018 EPFL
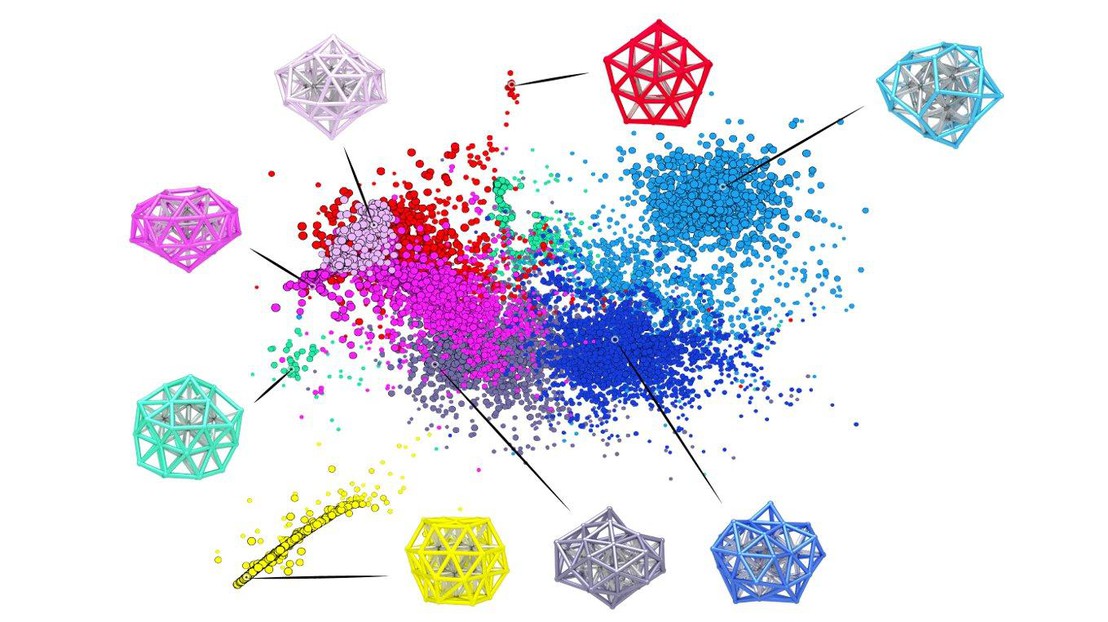
A new publication by the Laboratory of Computational Science and Modelling demonstrates how automatic pattern recognition can be used to identify the building blocks of nanoclusters and proteins.
Most of the current understanding of structure-property relations at the molecular and the supramolecular scales can be formulated in terms of the stability of and the interactions between a limited number of recurring structural motifs (e.g. H-bonds, coordination polyhedra, protein secondary-structure). In an article that has just been published in the Journal of Chemical Theory and Computation an algorithm is introduced to automatically recognize such patterns, based on the identification of local maxima in the probability distributions observed in atomistic computer simulations, which is robust to the dimensionality and the sparsity of the reference atomistic data. It can be applied to very different classes of systems, for instance to identify coordination environments in Lennard-Jones clusters, and to recognize secondary-structure patterns in the simulation of an oligopeptide. To assess the applicability of this algorithm for motifs that involve several interdependent degrees of freedom, it is also used to identify groups of conformers of the cluster and the polypeptide, considered in their entirety. The motifs identified by analyzing atomistic simulations can be used to interpret and rationalize the stability and behavior of the system at hand, and also as a tool to accelerate sampling, in association with biased molecular dynamics schemes.