New release of Blue Brain Project Atlas sheds light on neuron types
2023 EPFL/ Blue Brain project- CC-BY-SA 4.0
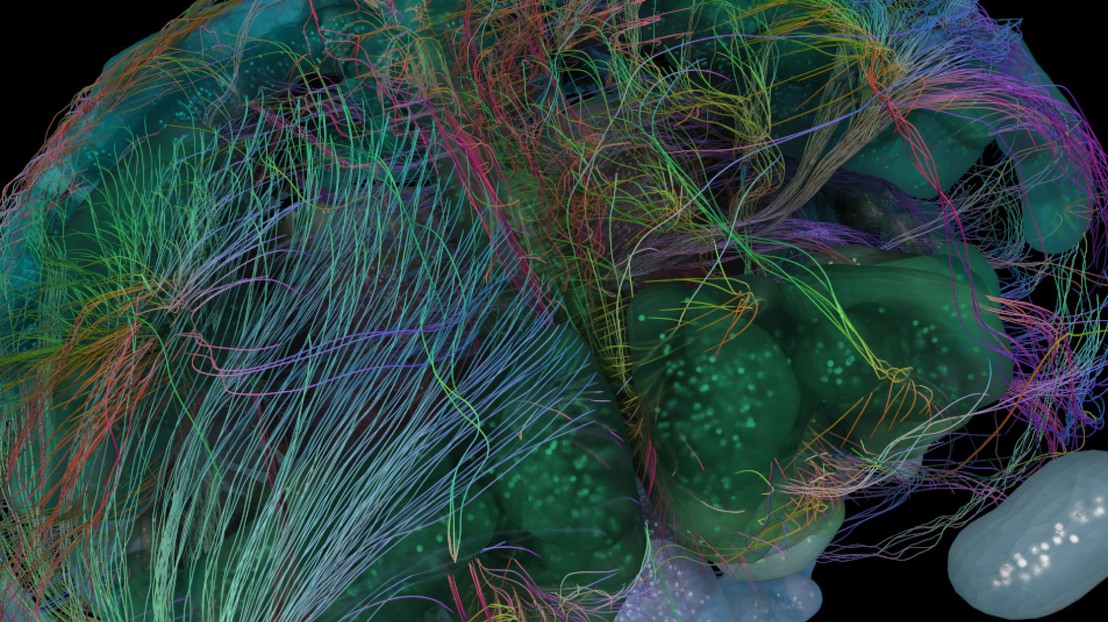
After four years of research, EPFL’s Blue Brain Project shares an enriched version of their 3D digital cell atlas of the mouse brain which includes more neuron types. The new approach can be extended to any other cell type, and provides a resource to build tissue-level models of the mouse brain.
Knowledge of the cell-type specific make-up of the brain is useful to understand the role of each cell type as part of the network, is necessary to tackle any large scale neural circuit simulation, and is key to Blue Brain's long term goal of accurately building a digital model of the whole mouse brain. Nonetheless, obtaining a global understanding of the cellular composition of the brain is an excessively complex task, not only because of the great variability inherent in the literature but also because of the numerous brain regions and cell types that make up the brain.
In 2018, EPFL’s Blue Brain Project presented the first model of a cell atlas which provided an estimate of the composition of the mouse brain. The release of Blue Brain’s Cell Atlas (BBCAv1) marked the first time a 3D digital atlas provided information on major cell types, numbers and positions in all the more than 700 regions of the mouse brain. It provides the densities of neurons, the associated connective tissue cells (glia) and their subtypes for each region, all of this presented in a navigable and dynamic format, allowing researchers to contribute new data. “At the time, it filled the huge gap in our knowledge of 96% of the mouse's brain regions,” says Blue Brain Founder and Director, Professor Henry Markram.
In recent years, new datasets and tools have emerged, providing cell-type composition based on the specific proteins expressed within the cells. While comparatively quick, these molecular marker techniques alone do not always yield directly usable information on the morphologies (shape) and electrophysiological properties of neurons. However, characterizing morpho-electrical properties of cells is extremely time-consuming and not suited to whole brain scans. It is therefore desirable to bring together and combine all the various available datasets in order to create one coherent framework with as much detailed information as possible.

2023 EPFL/ Blue Brain project- CC-BY-SA 4.0
Revealing inhibitory neuron density
One notable class of neurons for which very little data was available, and for which the method used to establish the BBCAv1 needed to be refined, is inhibitory neurons. Inhibitory neurons dampen the firing of other neurons and play a crucial role in packaging and transmitting information in the brain. They act like neuronal punctuation marks, and allow the brain to make sense of the influx of information. Estimates of inhibitory neuron counts were collected from the literature and a framework was built in order to combine them consistently into the cell atlas. Using brain slice images, inhibitory neuron densities were also estimated in regions where no literature data was available. In total, the authors reveal that in the mouse brain 20% of all neurons are inhibitory. “This sets the stage for subdividing inhibitory neurons into more fine-grained classes,” according to lead author, Blue Brain’s Dimitri Rodarie “and allows the neuroscience community to identify areas where current knowledge can be enhanced by additional constraints”.
Cross-species help for neuron models
The information mined from the Allen Institute for Brain Science provides essential data, allowing the creation of a catalog of neurons in the mouse brain according to their molecular, morphological and electrophysiological properties. However, in order to model brain regions, and more so a whole brain, not only is a global understanding of the cellular composition of the brain required, but detailed biophysical models of neurons must also be created. In a previous publication (Markram et al. 2015), Blue Brain built models based on morpho-electric data from neurons of the juvenile rat somatosensory cortex. As the data is from different species - mouse vs rat - and from a different developmental stage, the authors included normalization steps in order to map the models to the cell data from the Allen Institute. This step not only allowed them to assign a molecular identity to the neuron models, but also to populate the whole mouse cortex with detailed neuronal models. “Our algorithm helps to draw parallels across species but also extends our understanding of less studied brain areas”, explains lead author, Blue Brain’s Yann Roussel, adding “This model will allow experimentalists to understand regional composition and allow computational neuroscientists to place defined cell types in their simulations”.
The new tools and methods used to refine the Cell Atlas and produce the BBCAv2, published in two companion papers in PLOS Computational Biology, were extended to map well identified types to inhibitory neuron subclasses, paving the way for more accurate in silico reconstructions of brain tissues. The data, algorithms, software, and results of the pipeline used to upgrade the Blue Brain Cell Atlas are all publicly available. For Daniel Keller, leader of Blue Brain’s Molecular Systems team "This version encompasses four years of studies and includes additional constraints from biological data to make the results more amenable to simulation. Using it for simulation allows us to identify areas for further refinement, thereby permitting improvement with every successive generation.”
“This project aims to involve the scientific community to contribute with open access to data, software and tools. We expect the BBCAv2 to be used for many purposes,” conclude the authors.
This study was supported by funding to the Blue Brain Project, a research center of EPFL, from the Swiss government’s ETH Board of the Swiss Federal Institutes of Technology.
Roussel Y, Verasztó C, Rodarie D, Damart T, Reimann M, Ramaswamy S, et al. (2023) Mapping of morpho-electric features to molecular identity of cortical inhibitory neurons. PLoS Comput Biol 19(1): e1010058. https://doi.org/10.1371/journal.pcbi.1010058
Rodarie D, Verasztó C, Roussel Y, Reimann M, Keller D, Ramaswamy S, et al. (2022) A method to estimate the cellular composition of the mouse brain from heterogeneous datasets. PLoS Comput Biol 18(12): e1010739. https://doi.org/10.1371/journal.pcbi.1010739