How a highly unstable protein may lead to neurodegeneration
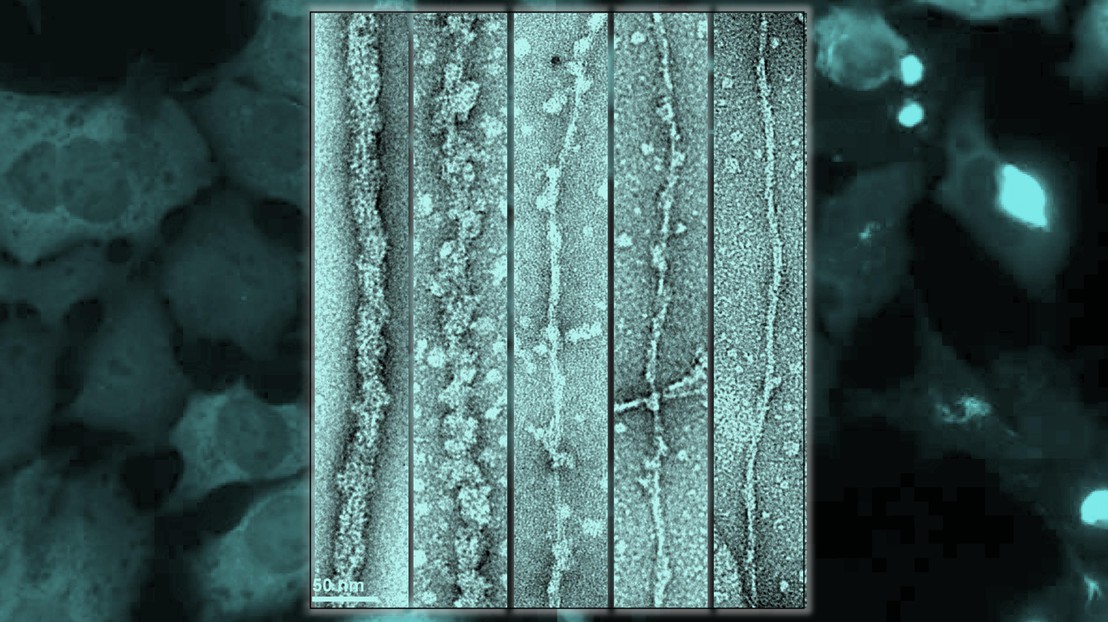
Illustration depicting how exposingTDP-43 fibril core enhances pathology formation in cell. 2023 EPFL / Galina Limorenko - CC-BY-SA 4.0
EPFL scientists reproduce key features of pathological protein aggregates found in the brain of patients with Lou Gehrig's disease and other neurological diseases, providing insight into the underlying mechanism and offering promising avenues for new therapies.
Several neurodegenerative diseases, such as Alzheimer’s, Parkinson’s, and Lou Gehrig’s Disease aka Amyotrophic lateral sclerosis (ALS), are caused by proteins that go stray and start to aggregate into fibrils that accumulate in specific brain regions. Now, EPFL scientists have discovered a new mechanism that explains how the aggregates become pathological and spread to different regions of the brain. A main suspect is the highly unstable protein called TDP43. The scientists discovered that TDP43 aggregates that form in the brain are not implicitly pathogenic until they are processed to reveal their “sticky” core. The results are published in Nature Neuroscience.
Aggregation of the protein TDP43 is a hallmark of ALS and other neurodegenerative diseases. Once formed, TDP43 aggregates can spread to different brain regions where they corrupt the normal and functional TDP-43. But what triggers TDP-43 aggregation in the first place? What are the mechanisms responsible for unleashing its pathogenic effects? This knowledge gap hinders the development of effective drugs to block TDP-43 aggregation or neutralize its toxic properties.

In the foreground, TDP-43 fibrils comprising full-length protein are being digested by the enzyme Proteinase K (left to right), thinning out the fibril by exposing the amyloid core. In the background, the cell model shows the seeding by the fibrils only when the core is exposed – on the right side, the bright blobs, whereas when the fibril core is masked, on the left, there is no seeding of the aggregates in cells, so we can see the diffuse staining only. This shows the amyloid core must be unmasked to make the fibrils toxic and seeding-competent.
Unleashing the pathogenic effects of TDP43 by cleavage
In this latest EPFL study led in collaboration with scientists from the University of Pennsylvania, Dr. Senthil Kumar and Prof. Hilal Lashuel discovered a new mechanism responsible for unleashing the pathogenic effects of TDP43 aggregate, prepared in the test tube or isolated from postmortem patients’ brains. The surfaces of these TDP43 aggregates must first be cleaved by enzymes to reveal hidden sticky surfaces which attract normal TDP-43 proteins and induce the formation of more aggregates.
“The discovery was facilitated by our ability to develop a new method for the production of fibrils, in the laboratory, that share morphological and structural features with those found in the brain of patients with ALS”, says Dr. Senthil T. Kumar, the first author of the paper.
Using cryo-electron microscopy, where samples are cryogenically frozen before being viewed through an electron microscope, the researchers showed that TDP-43 filaments are buried inside a larger filament and are inaccessible, i.e. not yet pathological, because they are covered by the globular parts of the protein. As long as these filaments are buried, they exist in stealth mode and are not accessible to other molecules or proteins. In other words, TDP43 becomes pathological when its outer coating is cleaved to reveal its “sticky”, inner filaments, yet remains in stealth mode when its outer coating is intact.
“Our findings suggest that inhibiting the enzymes responsible for cleaving the TDP-43 filament represents a viable therapeutic strategy to slow the formation of TDP-43 aggregates and prevent their spreading in the brain, thus slowing the progression of the disease. As a next step, we plan to identify these enzymes and determine whether inhibiting their activity could prevent TDP-43 aggregation and neurodegeneration in cell and animal models of ALS”, says Hilal Lashuel, EPFL professor who runs the lab that led the study.
The new results also have implications for developing new tools and methods for early diagnosis of ALS and other neurodegenerative diseases. The protective globular layer may explain why TDP-43 fibrils are so hard to detect. Standard methods and dyes commonly used to detect and monitor fibril formation by other protein suspects in the brain often failed to detect TDP-43 fibrils. “It also explains why it has been very challenging to develop imaging agents using intact TDP-43 fibrils. Such imaging agents are desperately needed to enable early diagnosis, monitor disease progression and assess the efficacy of new therapies”, says Dr. Kumar.
Importance of studying the full-length protein
TDP-43 is a highly unstable protein and quickly aggregates into different structures, thus making it challenging to generate pathology-resembling TDP-43 aggregates in a reproducible manner. This has forced many scientists to work with small fragments of the protein, in particular fragments from the region responsible for driving its aggregation. “When we determined the structure of the protein fragment that form the core of the TDP-43 fibrils prepared in the lab, we obtained a different structure than that of TDP-43 fibrils isolated from patients’ brain, even though the amino acid sequence of these fragments is virtually identical”, says Dr. Kumar.
“Our findings demonstrate that protein sequences flanking this aggregation-prone region play important roles in dictating the final structure and that reproducing the properties of TDP-43 aggregates in the brain requires working with the full-length protein”, says Hilal Lashuel. “This is essential to ensure that the drugs, antibodies, and imaging agents we develop in the laboratory will have higher chances of engaging the disease-relevant TDP-43 aggregates in the brain of patients.”
The researchers showed that they can produce TDP-43 fibrils with the same core sequence as that of fibrils from patients’ brains. “But we still have to determine if the unmasked fibril core has the same structure”, clarifies Hilal Lashuel. “If we show this, then we will have the only system that allows one to produce the actual pathology in the test tube. This will have huge implications for understanding how disease-linked mutations and protein modifications influence TDP-43 aggregation and would facilitate the development of new drugs that block TDP-43 aggregation, neutralize its pathogenicity or bind to TDP-43 aggregates and facilitate their detection in the brain.