First Digital Reconstruction of the Neuro-Glia-Vascular Architecture
© 2021 EPFL
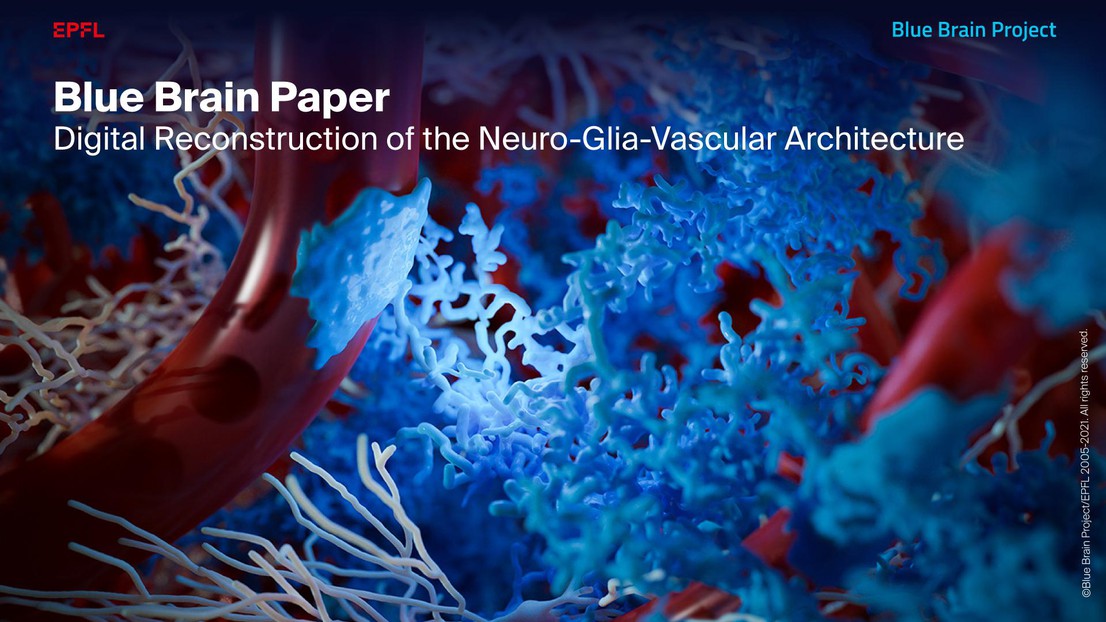
Blue Brain has created the first digital reconstruction of the Neuro-Glia-Vascular (NGV) Architecture providing a new framework to study brain function in health and disease.
The study, published in Cerebral Cortex, represents a major milestone for the EPFL Blue Brain Project because they can now reconstruct the architecture of non-neuronal entities such as blood vessels and the supporting cells called glia. This means it is possible to capture the way that neurons, glia and the blood supply interact. These reconstructions of the brain tissue provide a sub-micron precise framework needed to simulate the molecular interactions relevant to understanding how neurons are supported and nurtured. They can also be used to investigate how drugs interact and explore how neurodegenerative diseases arise. Blue Brain has made all the experimental data, models and tools used to reconstruct brain tissue at this resolution, open source in the Blue Brain NGV web portal.
As you read this article, your brain is engaging neurons and glia to process the information. This eventually increases the demand for neuronal metabolic energy leading to increased production of energy and local changes in the diameter of the proximal vessels to balance out this demand. The complex structural organization of the two dominant cell classes in the brain, neurons and glia, which play a role in these effects, is known as the Neuro-Glia-Vascular (NGV) system. Now, for the first time, Blue Brain scientists have digitally reconstructed the architecture of this system including neurons, glia and blood vessels.
“We created a data-driven digital reconstruction of the NGV architecture at sub-micrometer resolution using Blue Brain’s state-of-the-art model of neural microcircuitry (Markram et al., 2015). We combined the latter with literature and experimental data to reconstruct the spatial arrangement and morphologies of astrocytes (the most common glial type in the gray matter) and their relation to the vasculature in the rat cortex,” explains lead author, Eleftherios Zisis. “We digitally mapped the shape of glia using an electron microscope and then mathematically recreated their shapes. We then built a 0.2 mm3 of rat somatosensory cortical tissue with 16,000 morphologically detailed neurons, 2500 protoplasmic astrocytes, and its microvasculature. The consistency of our reconstruction with its wide array of experimental measurements allows novel predictions of the NGV organization. This allows anatomical reconstruction of overlapping astrocytic microdomains and quantification of end-feet (astrocytic terminals that wrap around blood vessels) connecting each astrocyte to the vasculature, as well as quantification of the extent to which they end-feet cover the blood vessels. This means we can look deeper into how the NGV system is organized and how it works. We were able to perform simultaneous measurements of both geometrical and functional aspects of the NGV, which is often difficult to perform with experimental setups,” he concludes.
Interdependencies and topological methods allow dense in silico reconstruction from sparse experimental data
Sparse experimental data is an on-going problem for science and the experimental reconstruction of astrocytes is particularly difficult and expensive; therefore, the community cannot easily obtain the numbers of cells required for creating full circuits - circuits that may require from thousands to millions of cells, depending on the size of the brain region to reconstruct. The Blue Brain has already developed a novel mathematical approach using topology to describe the branching structure of neurons (Kanari et al) and we applied this approach to astrocytes that have much finer branches to create thousands of unique astrocytic morphologies for each astrocytic domain, thus solving the problem of insufficient experimental data.
“By assembling the statistics of neural tissue, a coherent picture emerges of how astrocytes are organized in the brain,” states Daniel Keller, leader of Blue Brain’s Molecular Systems team. “We hope that sharing our findings will motivate experimentalists to understand what types of data are most useful for their approach and will provide a framework for how new data can be incorporated. To ensure biological fidelity, the NGV circuit was validated against values reported in anatomical studies and experimental measurements at different levels of detail, ranging from how many astrocytes there are down to the structure of single astrocytes.”
In silico approach minimizes costs and time of scientific discovery
The Blue Brain in silico anatomical reconstructions of the NGV do not seek to replace experimental measurements but to minimize the costs and time required for scientific discoveries. Algorithmically generated NGV circuits can serve as a magnifying glass into the brain's complexity, allowing scientists to explore the geometry and topology of its cells and their connections. Moreover, the creation of multiple NGV circuits, each one with a different set of parameters that reflect organizational changes in brain anatomy, could allow for a better understanding of the anatomical principles and their geometric constraints. With these insights, scientists can develop more focused experiments, saving time and costs.
Furthermore, the study showed that an in silico circuit of the NGV architecture could simultaneously quantify both compositional (densities, wiring, surface areas, and volume) and organizational (connectivities, distance distributions, correlations) aspects of its entities, as well as investigating questions concerning the intricacies of cell type composition and their relation to computational capacity.
Facilitator for research into drug delivery and neurodegenerative diseases
“One needs a very high resolution view of the anatomy of the NGV system to begin understanding pathologies and what treatments could be developed,” states Blue Brain Founder and Director, Prof. Henry Markram. “Drug delivery research studies the interactions between drugs and molecules and how molecules interact with each other in constrained spaces. This Blue Brain model provides a structural foundation for biophysical models to explore cross-talk between neurons, glia, and the vasculature at a resolution that was not possible before”.
Similarly, research in neurodegenerative diseases such as Alzheimer's disease targets reactive astrocytes, the morphology of which is entirely transformed with variation in their branching, overlap, and proliferation compared to healthy brains. For both cases, this data-driven approach allows for incremental improvements as more experimental data become available, as new biophysical models are published, and as new questions arise,” he summarizes.
Using the reconstruction of the structural foundations of the NGV, the next step for Blue Brain is to simulate metabolic processes occurring within the NGV ensemble, as well as modulation of blood flow mediated by NGV components. All of which will be freely available via the Blue Brain NGV portal.
-----------------------------
For more information, please contact Blue Brain Communications Manager – [email protected]
Citation - Zisis, E., Keller, D., Kanari, L., Arnaudon, A., Gevaert, M., Delemontex, T., Coste, B., Foni, A., Abdellah, M., Calì, C., Hess, K., Magistretti, P. J., Schürmann, F., & Markram, H. (2021). Digital reconstruction of the neuro-glia-vascular architecture. Cerebral Cortex. Volume 31, Issue 12, December 2021, Pages 5686–5703, https://doi.org/10.1093/cercor/bhab254
NGV Portal
The data that support the findings of this study (experimental data, models and tools used in the circuit building process) are openly available in the NGV portal website at Blue Brain NGV portal.
Funding
This study was supported by funding to the Blue Brain Project, a research center of the École polytechnique fédérale de Lausanne (EPFL), from the Swiss government’s ETH Board of the Swiss Federal Institutes of Technology, and is based upon work supported by the King Abdullah University of Science and Technology (KAUST) Office of Sponsored Research (OSR) under Award No. OSR-2017-CRG6-3438.
Acknowledgements
This work was developed as part of the King Abdullah University of Science and Technology (KAUST) and EPFL Blue Brain Project Alliance for Integrative Modeling of Brain Energy Metabolism. We are also grateful to Bruno Weber of ETH Zurich for providing a previously published vasculature data set.
About the EPFL Blue Brain Project
The aim of the EPFL Blue Brain Project, a Swiss brain research initiative founded and directed by Professor Henry Markram, is to establish simulation neuroscience as a complementary approach alongside experimental, theoretical and clinical neuroscience to understanding the brain, by building the world’s first biologically detailed digital reconstructions and simulations of the mouse brain. https://www.epfl.ch/research/domains/bluebrain/
About EPFL
EPFL, one of the two Swiss Federal Institutes of Technology, based in Lausanne, is Europe’s most cosmopolitan technical university with students, professors and staff from over 120 nations. A dynamic environment, open to Switzerland and the world, EPFL is centered on its three missions: teaching, research and technology transfer. EPFL works together with an extensive network of partners including other universities and institutes of technology, developing and emerging countries, secondary schools and colleges, industry and economy, political circles and the general public, to bring about real impact for society. epfl.ch