Discovery of a new mechanism for bacterial division
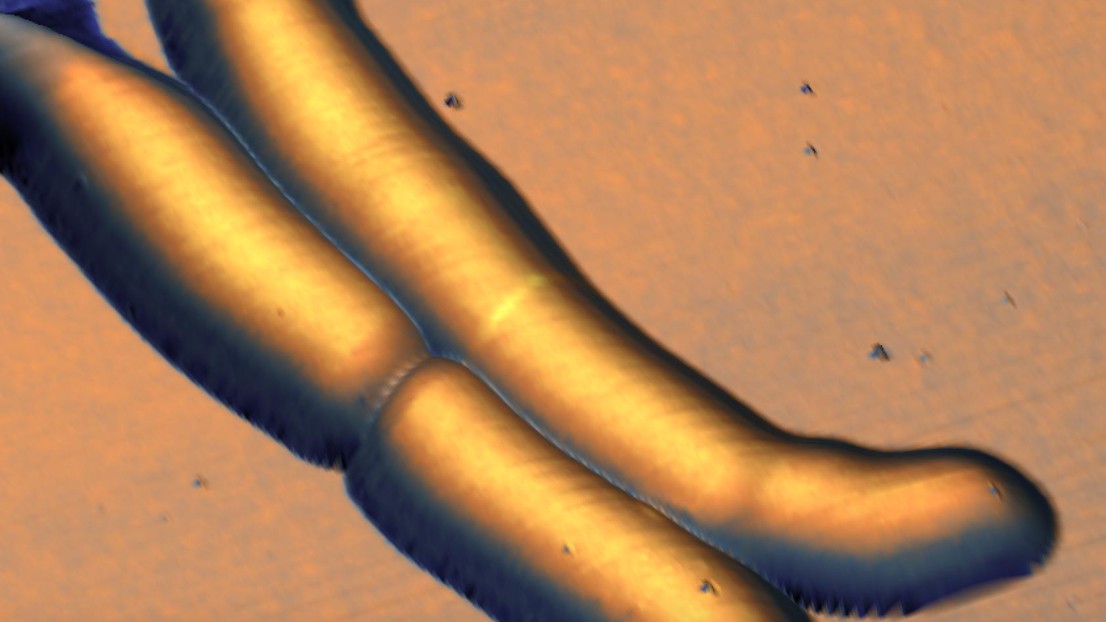
The wave-trough division site in M. smegmatis as seen with the techniques developed in this study © J. McKinney/G. Fantner/EPFL
EPFL scientists show how some pathogenic bacteria — such as the mycobacteria that cause tuberculosis — use a previously unknown mechanism to coordinate their division. The discovery could help develop new ways to fight them.
Most rod-shaped bacteria divide by splitting into two around the middle after their DNA has replicated safely and segregated to opposite ends of the cell. This seemingly simple process actually demands tight and precise coordination, which is achieved through two biological systems: nucleoid occlusion, which protects the cell’s genetic material from dividing until it replicates and segregates, and the “minicell” system, which localizes the site of division around the middle of the cell, where a dividing wall will form to split it in two. But some pathogenic bacteria, e.g. Mycobacterium tuberculosis, don’t use these mechanisms. EPFL scientists have now combined optical and atomic force microscopy to track division in such bacteria for the first time, and have discovered that they use instead an undulating “wave-pattern” along their length to mark future sites of division. The findings are published in Nature Microbiology.
The work was carried out jointly by the labs of John McKinney and Georg Fantner at EPFL. The scientists wanted to understand how bacteria that do not have the genes for nucleoid occlusion and the minicell system “decide” where and when to divide. This is important, as many pathogenic bacteria fall into this category, and knowing how they divide can open up new ways to fight them.
The researchers focused on Mycobacterium smegmatis, a non-pathogenic relative of M. tuberculosis. Neither of these bacteria uses the two “conventional” biological systems for coordinating division, meaning that a non-conventional approach was needed for studying them.
The researchers combined two types of microscopy to track the life cycle of the bacteria. The first technique was optical microscopy, which uses fluorescent labels for “seeing” various biological structures and biomolecules. The second technique was atomic force microscopy, which provides extremely high-resolution images of structures on the cell surface by “feeling” the surface with a tiny mechanical probe, much like a blind person can form a three-dimensional mental image of an object by passing their hands over its surface.
“This experiment constitutes the longest continuous atomic force microscopy experiment ever performed on growing cells,” says Georg Fantner, while John McKinney adds: “It illustrates the power of new technologies not only to analyze the things we already knew about with greater resolution, but also to discover new things that we hadn’t anticipated.”
Armed with a custom-built instrument that combines the two techniques, the scientists created long-term time-lapses of the growth and division of the bacteria over multiple generations. Unexpectedly, they found that the bacteria produce undulating “trough-like” patterns across their length. These morphological landmarks on the undulating surface of mycobacterial cells correspond to future sites of cell division.
The troughs are roughly repeating waves, which the scientists calculated to have an average wavelength of ~1.8 μm, and an amplitude too small to resolve with conventional microscopes (about 100 nm). This might be the reason why waveform troughs have not been reported before.
The time-lapse images also showed that, after the mycobacterium divides, the new “daughter” cells inherit the “mother” cell’s wave-trough pattern, and ultimately divide at the center-most wave-trough.
Wave-troughs can form up to three generations before they are used as division sites. According to Alexander Eskandarian, the lead author of the study, these morphological features are “by far the earliest known landmark of future division sites in bacteria." Building on these observations, future research will focus on identifying the underlying mechanisms responsible for wave-trough formation and propagation and recruitment of the cell division machinery.
This work was funded by the Swiss National Science Foundation (SNSF), the Innovative Medicines Initiative, the European Union Seventh Framework Programme (EU-FP7), and the European Molecular Biology Organization (EMBO).
Reference
Haig A. Eskandarian, Pascal D. Odermatt, Joëlle X. Y. Ven, Mélanie T. M. Hannebelle, Adrian P. Nievergelt, Neeraj Dhar, John D. McKinney, Georg E. Fantner. Division site selection linked to inherited cell-surface wave-troughs in mycobacteria.Nature Microbiology 26 June 2017. DOI: 10.1038/nmicrobiol.2017.94