Dimitris N. Chorafas Foundation Award 2015 – Mingfu Shao

© 2015 EPFL
Models and Algorithms for Comparative Genomics
Thesis director: Prof. B. Moret
“For innovative formulations of integer-linear programs and creative uses of preprocessing and constraint generation for solving such integer-linear programs efficiently in the study of genome evolution through duplications, losses, and rearrangements.”
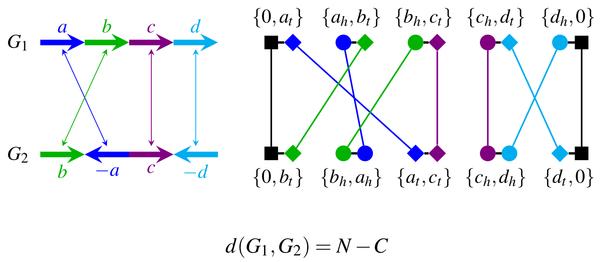
We develop novel algorithms for the fundamental computational problems in whole-genome comparison, i.e., to compute the edit distance (for two genomes) or median distance (for multiple genomes). For pairwise whole-genome comparison, we designed the first exact algorithm to compute the edit distance under the DCJ (double-cut-and-join) model, and the first exact algorithm to compute the edit distance under DCJ operations and segmental duplications, both in the presence of duplicate genes. We devised a (1.5 + epsilon)-approximation algorithm to compute the edit distance under DCJ operations, insertions, and deletions. We also proposed a very fast and exact algorithm to compute the exemplar breakpoint distance. For multiple whole-genome comparison, we designed a polynomial-time algorithm using a network flow formulation to compute the so called adequate subgraphs--a central phase in computing the median. We also proved that an existing upper bound of the median distance is tight.
We also apply our algorithms to systematically infer functional relationships and annotate genomes. For example, we applied our methods to infer orthologs and in-paralogs between a pair of genomes. On biological whole-genome datasets, our methods run very fast, scale up to whole genomes, and also achieve very high accuracy.