Automatic Selection of Descriptors for Machine Learning of Materials
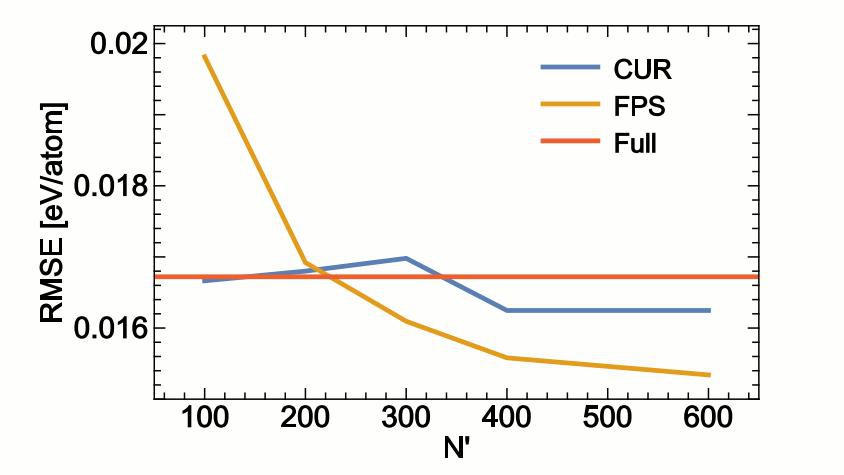
ML Accuracy as a function of descriptor size © COSMO / 2018 EPFL
What is the most effective representation of atomistic models to be used as the input for machine learning? A paper from the Laboratory of Computational Science and Modelling demonstrates how to let an algorithm make the optimal choice.
Machine learning of atomic-scale properties is revolutionizing molecular modeling, making it possible to evaluate inter-atomic potentials with first-principles accuracy, at a fraction of the costs.The accuracy, speed, and reliability of machine learning potentials, however, depend strongly on the way atomic configurations are represented, i.e., the choice of descriptors used as input for the machine learning method. The raw Cartesian coordinates are typically transformed in “fingerprints,” or “symmetry functions,” that are designed to encode, in addition to the structure, important properties of the potential energy surface like its invariances with respect to rotation, translation, and permutation of like atoms. A recent paper from the Laboratory of Computational Science and Modelling discusses automatic protocols to select a number of fingerprints out of a large pool of candidates, based on the correlations that are intrinsic to the training data. This procedure can greatly simplify the construction of neural network potentials that strike the best balance between accuracy and computational efficiency and has the potential to accelerate by orders of magnitude the evaluation of Gaussian approximation potentials based on the smooth overlap of atomic positions kernel. Examples of its application to the construction of neural network potentials for water and for an Al–Mg–Si alloy and to the prediction of the formation energies of small organic molecules using Gaussian process regression demonstrate the potential of this approach.