An enzyme “family business” holds the key to pluripotency
© Daniel Constam/EPFL
EPFL scientists have identified three enzymes that jointly regulate the formation of pluripotent stem cells.
Early on in mammalian fetal development, a few stem cells that will form all tissues of the fetus are set aside from an outer shell of so-called trophectoderm cells. However, we know very little about what regulates this initial cell differentiation. In a breakthrough study, EPFL scientists have identified three enzymes that control this differentiation by cooperating in the way they process the protein E-cadherin. The work is published in the Journal of Cell Biology.
When an egg is fertilized, it divides into two cells, then four, and so forth. When it reaches eight cells, the structure is called the “morula”. At this point, cell-cell contacts begin to accumulate E-cadherin molecules, which stick to each other like velcro. This modifies the shapes of cells in a process called "morula compaction”.
During the next division, cells that become buried on the inside of the morula have more surface covered by their immediate neighbors than cells that are pushed to the outside. As a result, E-cadherin activates a molecular cascade known as the Hippo signaling pathway in the inner cells, while the same pathway is inhibited in differentiating outer cells.
The Hippo pathway can “sense” the crowdedness of cells in many different contexts e.g. to limit the final size of developing organs. However, in the early embryo, E-cadherin activates it to ensure that inner cells remain pluripotent. Despite multiple efforts, the question remains: how are morula compaction and the formation of the inner cell mass initiated?
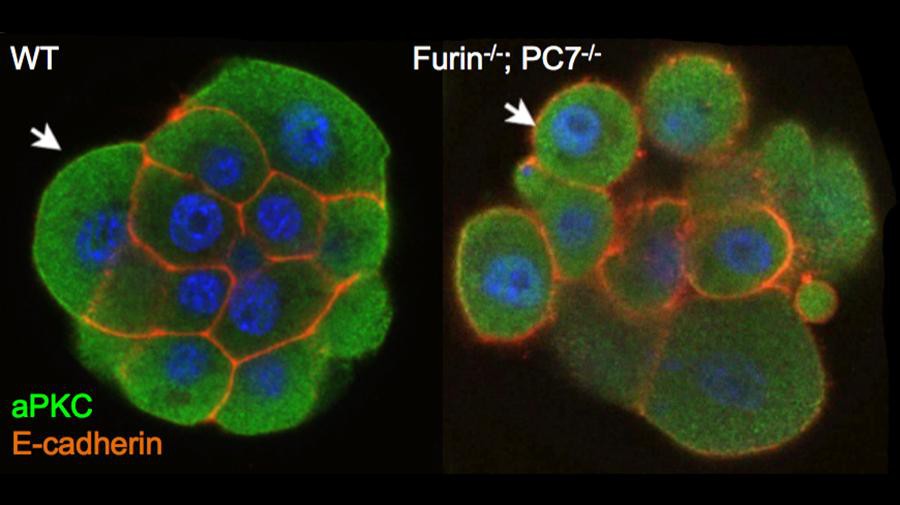
The answer seems to lie in the collaboration of three closely related proprotein convertases: Furin, PC7, and Pace4. The findings of Daniel Constam’s lab at EPFL show that all three enzymes contribute to a cleavage within the E-cadherin protein that activates its velcro-like adhesive function. To determine how this cleavage is regulated in mouse embryos, the Constam lab used a sensor called CLIP (Cell-Linked Indicator of Proteolysis), which they developed in 2010 to live-image when and where a cell’s convertases are active.
By using CLIP in combination with knockout mice lacking one, two or three proprotein convertases, the researchers were able to map the distribution of the enzymes that process E-cadherin in the compacted morula over time. The results showed that combined loss of Furin and PC7 inhibits CLIP activation and morula compaction, meaning that E-cadherin processing and activation initially depends on these two enzymes.
However, after an initial delay, compaction was partly rescued by the 'youngest cousine' Pace4, coinciding with the appearance of its processing activity. On the other hand, removing Furin and Pace4 revealed that development continued almost until gastrulation - only with PC7. This means that, just one member of this enzyme family can “fill in” if the other two are missing.
In order to map the activity and distribution of the enzymes in space, the researchers used a pharmacological inhibitor of proprotein convertases. As a result, they found striking differences between the localization of each enzyme, although all were found to be active in both inner and outer cells. Specifically, Furin was blocked in both outer and inner cells, PC7 in none, and Pace4 only in the inner cells.
The work reveals a new, joint role for the three enzymes, and casts light into the way that fetal development is controlled at a crucial juncture. “This is a remarkable redundancy built into the system, apparently to boost the robustness of its self-regulating properties,” says Daniel Constam. “It ensures that even if one of the three controls is missing, the fetus can still develop properly, and not all is lost.”
This work was funded by the Swiss National Science Foundation.
Reference
Bessonnard S, Mesnard D, Constam DB. PC7 and the related proteases Furin and Pace4 regulate E-cadherin function during blastocyst formation.Journal of Cell Biology 210(7):1185-1197. DOI: 10.1083/jcb.201503042