A new understanding of bacterial division
C. crescentus © 2014 Seamus Holden/EPFL
Scientists at EPFL have used a super-resolution microscopy technique to understand how bacteria divide. Their findings refute conventional models and can pave the way for the development of new antibiotics.
Bacteria are ubiquitous, and although 99% of them are harmless, the remaining 1% can cause wide-spread epidemics. In addition, our overt use of antibiotics promotes the appearance of resistant strains, creating an urgent need for new drugs. Now, a research team at EPFL has used cutting-edge microscopy to unravel the way bacteria reproduce. Their findings, published in PNAS, offer insights into a new target for the next generation of antibiotics.
In order to address diseases caused by bacteria, we need to understand how they function so that we can identify better ways of killing them. One tool at our disposal is fluorescent microscopy, where biological molecules are labelled with light-emitting “tags”. However, microscopes are limited in terms of resolution. The reason is that light behaves like a wave when it goes through obstacles like a microscope’s lenses and circular apertures. This phenomenon, called “light diffraction”, means that objects smaller than 200 nanometers appear blurry with a conventional microscope – a serious problem given that many biological molecules of interest are tens to hundreds of times smaller than that.
One way around the diffraction limit is photoactivated localization microscopy (PALM). Like fluorescent microscopy, PALM uses light-emitting (“photoswitchable”) tags, which are used to label proteins of interest in a cell, e.g. a bacterium. A few tags are then switched on, their signal in the cell is read and then they are deactivated to go “dark”. A different group of tags is then switched on and the process is repeated until a complete picture emerges. In this way, PALM can allow us to “see” at resolutions down to 10 nm, opening a whole new dimension of molecular visualization.
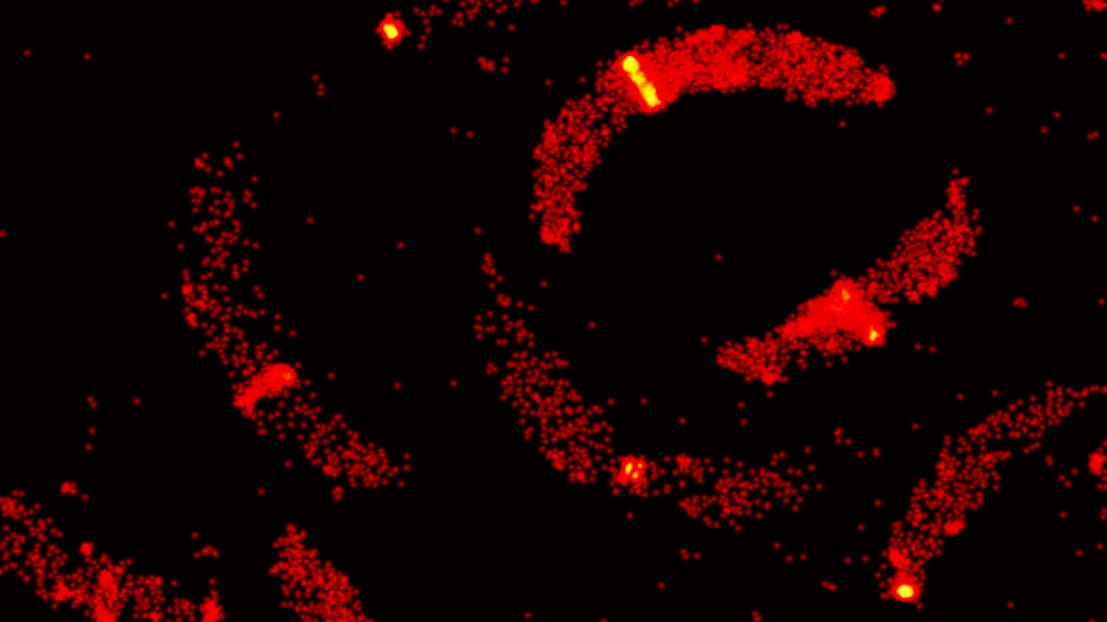
The team of Suliana Manley at EPFL has used a modified a 3D version of PALM to understand how bacteria divide. 3D PALM uses a weak cylindrical lens in addition to its regular spherical ones, which allows visualization on the z axis by creating an optical astigmatism effect. The researchers focused on a bacterial protein called FtsZ, which is part of the cytoskeleton of the vast majority of bacterial cells. Since its discovery, FtsZ has been thought to play a significant role in the division of bacteria by forming a continuous ring around the cell like a lasso, and constricting it to split it into two daughter cells. The structure, called a Z-ring, has made FtsZ a prime target for next-generation antibiotics.
Using a synchronized Caulobacter crescentus culture as a model, first author Seamus Holden designed a methodology that would allow him to visualize the division of individual C. crescentus bacteria with a custom-built 3D PALM system that he constructed over a year. The analysis showed that instead of forming a continuous ring, FtsZ instead appeared to form a “patchy midcell band” for the majority of the cell division process, with the ring forming only towards the end and in a minority of observed cells.
The data show for the first time how the Z-ring is organized in vivo, but alter the conventional model. In this new paradigm, FtsZ filaments are organized randomly across the midcell band, without clear evidence that they exert force to constrict the cell. Rather, the researchers believe that FtsZ might play more of an indirect role, recruiting other proteins that promote cell division in the bacterium. This new understanding of bacteria reproduction is expected to significantly impact the direction of antibiotic development in the future.
Reference
Holden SJ, Pengo T, Meibom KL, Fernandez CF, Collier J, Manley S. High throughput 3D super-resolution microscopy reveals Caulobacter crescentus in vivo Z-ring organization. PNAS doi:10.1073/pnas.1313368111