A new data format to efficiently describe large-scale network models

© 2020 EPFL
The increase in the availability of comprehensive experimental datasets and of high-performance computing resources are driving rapid growth in the scale, complexity, and biological realism of computational models in neuroscience. Accordingly, to support construction and simulation, as well as the sharing of such large-scale models, a broadly applicable, flexible, and high-performance data format is necessary.
To address this need, a global collaboration between the EPFL Blue Brain Project, the Allen Institute for Brain Science, the Paris-Saclay Institute of Neuroscience, the State University of New York, University College London and Stanford University has seen the development of the Scalable Open Network Architecture TemplAte (SONATA) data format. The design and architecture of SONATA builds on both the Blue Brain and Allen Institute’s expertise with large-scale high-performance network simulation, visualization and analysis.
The SONATA data format represents neuronal circuits and simulation inputs and outputs via standardized files and provides abundant flexibility for adding new conventions or extensions. The format has been designed for memory and computational efficiency and works across multiple platforms. SONATA can be used within multiple modeling and visualization tools, and reference Application Programming Interfaces and model examples to catalyze further adoption are also available.
Advancing our understanding of brain function and mechanisms
Jean-Denis Courcol, Blue Brain’s Neuroscientific Software Engineering Section Manager explains, “We developed SONATA with our collaborators at the Allen Institute to facilitate the exchange of our large-scale cortical models. Even though Blue Brain uses different modelling approaches and different tools than the Institute, the key element is that the format allows networks built by one party to be simulated by the other and vice versa. In doing so, we have gone some way to answering some of the challenges of modern computational neuroscience, especially those inherent in large-scale data-driven modelling of brain networks.”
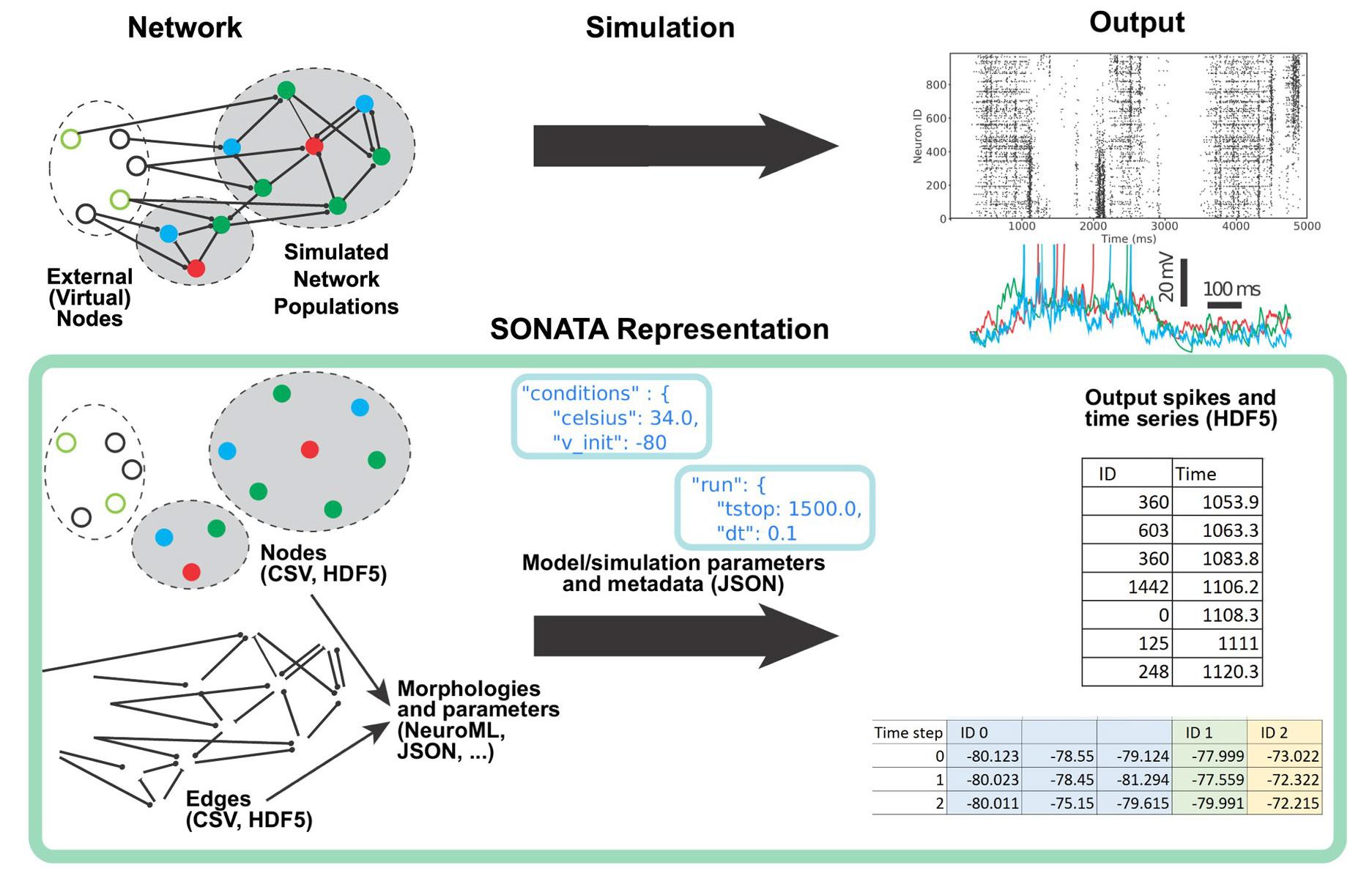
Anton Arkhipov, an Associate Investigator at the Allen Institute, says, “SONATA will help researchers to manipulate and share models of brain circuits efficiently, which is important for enhancing the scientific impact of such models. This collaboration between the Allen Institute, Blue Brain, and several other teams has been critical in producing a broadly applicable data format that will be useful for a wide spectrum of applications.”
SONATA is free and open for the neuroscience community to use and build upon with the goal of enabling efficient model building, sharing, and reproducibility. It already has support from several popular tools for model building, simulations, and visualization. Additionally, although SONATA has been originally developed to support very large and biologically complex simulations, it is fully consistent with more typical smaller-scale and less complex applications.
Nine authors from the Blue Brain were part of this broad collaborative effort that developed the SONATA data format for large-scale network models in a paper published in PLOS Computational Biology.
Dai K, Hernando J, Billeh YN, Gratiy SL, Planas J, Davison AP, et al. (2020) The SONATA data format for efficient description of large-scale network models. PLoS Comput Biol 16(2): e1007696. https://doi.org/10.1371/journal.pcbi.1007696.
All data and software are available at https://github.com/AllenInstitute/sonata and https://portal.brain-map.org/explore/models/l4-mv1.