A computational model describes cell motion
EPFL scientists discover how cells begin to move, and produce one of the most accurate computer models of cell movement.
In order to survive, cells have to move around. This begins with a process called cell polarization, wherein the cell reorganizes its cytoskeleton, signals, organelles, and membrane to change shape. Although it sounds simple, cell polarization is a fundamental question for cell biologists and physicists who have long attempted to describe the dynamics of cell motion. In a study published in Nature Physics, EPFL scientists have now discovered the novel and simple principle behind cell polarization, and have used it to develop an accurate computer model of cell movement.
An especially challenging question is the cell’s behavior before the onset of motion. We know that the cell undergoes a process known as “symmetry breaking”, where it abandons it’s natural symmetrical shape in preparation for movement. Symmetry breaking may be triggered by external stimuli, but can also happen spontaneously: the cell “chooses” the direction to go, starts moving, and then keeps going in the same direction. When moving, the cell “pushes” its front (called the “leading edge”) and pulls its back (called the “trailing edge”).
Models of cell movement aim to describe the polarization of this edge activity – in other words, how the cell finds its leading and trailing edges. They generally use gradients of regulatory proteins, and can also take into account feedback from the cell’s motion itself.
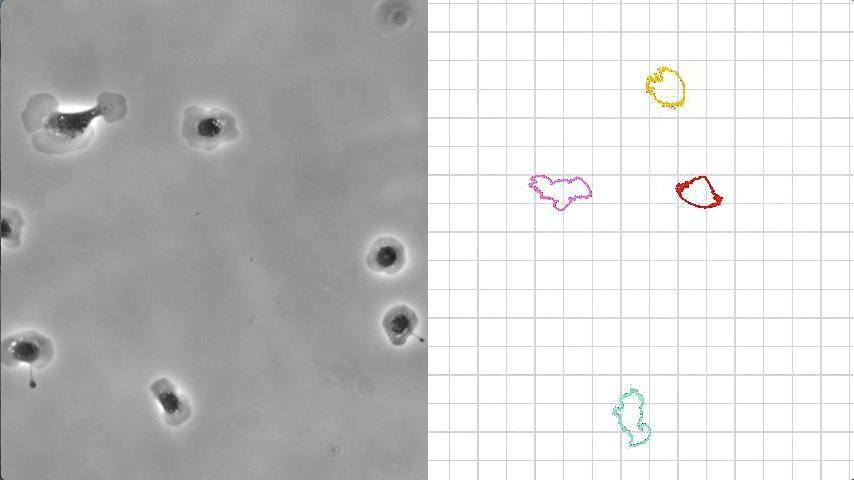
A research team led by Alexander Verkhovsky at EPFL used fish epidermal cells, called keratocytes, to investigate the dynamics of cell polarization. Keratocytes are a classic model for polarization and directional migration because they feature robust polarity, simple shape, and, helpfully, are constantly moving.
The scientists began by quantifying experimental data from observing cell-edge dynamics. This way, they could identify common features between cell polarization and movement.
The team found that the cell edge switches from protrusion to retraction through sensing the distance of the edge from the cell’s center. This ability to react to the distance is the same around the entire cell edge and in all directions, but, surprisingly, it helps the cell to choose its direction and separate its edge into leading and trailing parts. And even though a popular theory is that the cell has a “compass” – something that controls its local activity depending on the orientation with respect to its front–back axis – the researchers found no evidence to support it.
The team then used the experimental data to develop a computer model. “The idea was to demonstrate that distance-sensing is sufficient for spontaneous polarization and directional motion,” says Alexander Verkhovsky.
The model accurately reproduced the observed experimental data. Using stochastic algorithms, it was possible to faithfully reproduce a wide range of cell-migration behaviors. “I am very happy about this work,” says Verkhovsky. “Among other things, it shows that there are more ways to investigate cell behavior in 21st century than just trying to decipher a network of tens of thousands components; rather, simple and general concepts can still be discovered and drive research.” The group’s next goal is to uncover the physical and biological mechanisms behind distance-sensing, and to determine how common it is among different cell types and migration behaviors.
This work included a collaboration of EPFL’s Laboratory of Cell Biophysics with TU Dresden and the Max Planck Institute of Molecular Cell Biology and Genetics. It was funded by the Swiss National Science Foundation, the Swiss National Competence Center for Biomedical Imaging.
Reference
Raynaud F, Ambühl ME, Gabella C, Bornert A, Sbalzarini IF, Meister J-J, Verkhovsky AB. Minimal model for spontaneous cell polarization and edge activity in oscillating, rotating and migrating cells.Nature Physics 11 January 2016. DOI: 10.1038/NPHYS3615