A Bandpass Filter for Synthetic Biology
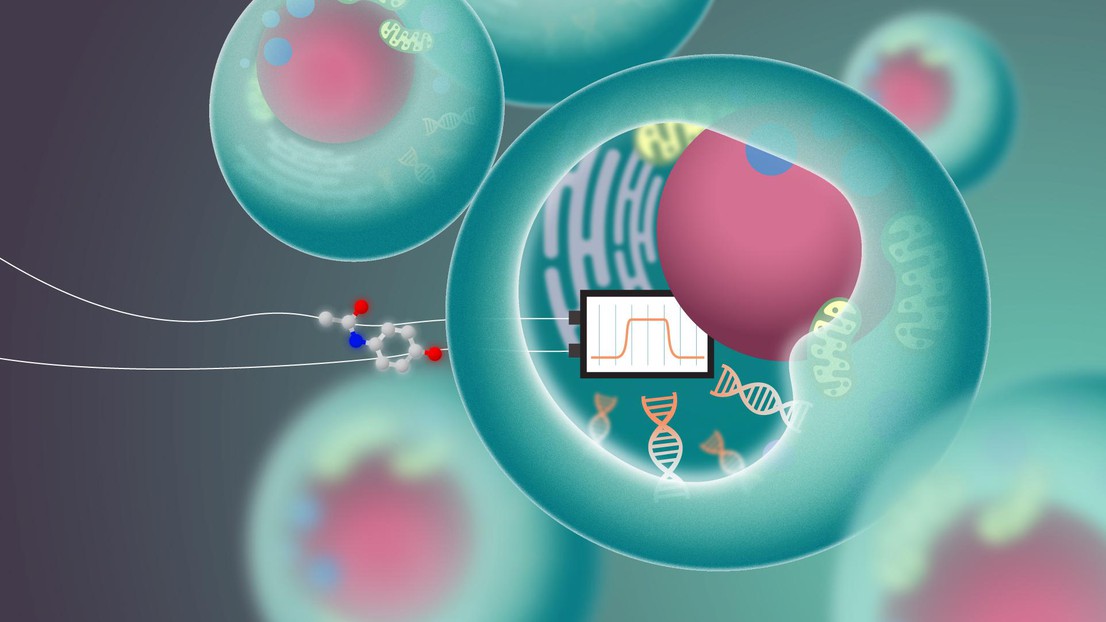
An illustration showing the concept of implementing a bandpass filter within a cellular environment. © 2023 EPFL/Titouan Veuillet - CC-BY-SA 4.0
EPFL scientists have crafted a biological system that mimics an electronic bandpass filter, a novel sensor that could revolutionize self-regulated biological mechanisms in synthetic biology.
Synthetic biology holds the promise of enhancing and modifying biological systems into innumerable new technologies for the benefit of society. This engineering approach to biology has already reaped benefits in the fields of drug delivery, agriculture, and energy production. In a paper published in Nature Chemical Biology, EPFL researchers at the Laboratory of Protein Design and Immunoengineering (LPDI). at the School of Engineering have taken an important step in designing more performative biological systems. By observing complex self-regulating behavior in cells and taking inspiration from electrical engineering, they have engineered a sophisticated biological switch using modified proteins as the hardware.
We use a number of approaches in terms of protein design, more specifically computational protein design, in order to engineer new molecular hardware.
The goal of LPDI is to develop biological functions that imitate and even go beyond what nature has provided over millions of years of evolution. “We use a number of approaches in terms of protein design, more specifically computational protein design, in order to engineer new molecular hardware,” says Bruno Correia, head of LPDI and lead author the paper. This new biological hardware—in the form of engineered proteins—can then be placed in living cells and respond to stimuli that the engineer can closely control.
The lab’s methodology begins by observing how cells function and imitating or adapting these functions for other uses. One of the things that nature does extremely well is self-regulation. When a cell needs more ions, for example, it activates mechanisms that allow for the ions to flow into the cell. Once the cell’s needs are met and it has reached a certain equilibrium known as homeostasis, it can then turn off the ion flow.
This biological mechanism can be conceptually likened to the selective nature of a bandpass filter in electronics. While a bandpass filter discriminates signals based on a specific range of frequencies, allowing only those within a designated band to pass through, the cell selectively permits ions to enter or exit based on its current needs. Although the comparison is a simplification, as the biological process is governed by complex biochemical feedback rather than binary signals, it similarly achieves selective permeability—akin to how a bandpass filter selectively permits certain frequencies while excluding others.
Sailan Shui took inspiration from the functioning of a bandpass filter and engineered its biological equivalent.
A current limitation in synthetic biology is that no design, up until now, offers this type of functionality—it’s all or nothing, only ‘on’ or only ‘off’. Think of penicillin delivery. As there is no regulatory system based on biological sensors for drug dosing, a diabetic needs to continuously monitor the insulin level. Sailan Shui’s work at LPDI focused on these types of biological sensors and switches that could drastically improve drug delivery and other biological systems. “Shui was unsatisfied by the on/off paradigm and decided to engineer a switch that could respond to changes in a cell’s internal and external environment. So she took inspiration from the functioning of a bandpass filter and engineered its biological equivalent,” explains Correia.
To create this novel function in biological systems, the team designed proteins and inserted them into living cells. In biology, function flows from form. Correia and Shui observed the structure of folded proteins and their effect on self-regulating functions inside the cell. They then created computational models that could potentially act as a bandpass filter from these observations. Once the digital design was validated using computer simulations, they started working on building the protein structure by manipulating its DNA and amino acid configuration. Finally, they tested the design in cell cultures. The results are conclusive. Their design, which they share with the research community in the spirit of open research, will most likely be used by researchers around the world in ways yet foreseen.
As a hypothetical application in synthetic biology, researchers could create a system that operates similarly to an electronic bandpass filter to regulate insulin delivery based on blood glucose levels. Engineered proteins would serve as sensors, detecting high glucose and triggering insulin release until levels normalize. This would automate insulin dosing, potentially improving diabetes management and reducing the need for frequent monitoring. Such a system would represent a significant advancement in the use of synthetic biology for therapeutic applications.
This fundamental research is essential for developing the tools, building blocks and hardware of the future of synthetic biology. “We have a clear methodology but also embrace the serendipity of science. It was the scientist Leo Scheller in my lab who had the vision to understand the importance of this work. It was a team effort,” says Correia. A team effort that brings the field one important step closer to engineering better drug delivery technology, more efficient bioreactors, and even entirely new forms of biological entities.
Shui, S., Scheller, L. & Correia, B.E. Protein-based bandpass filters for controlling cellular signaling with chemical inputs. Nat Chem Biol (2023). https://doi.org/10.1038/s41589-023-01463-7