Synthetic receptors can rewire cell functions and reduce side-effects
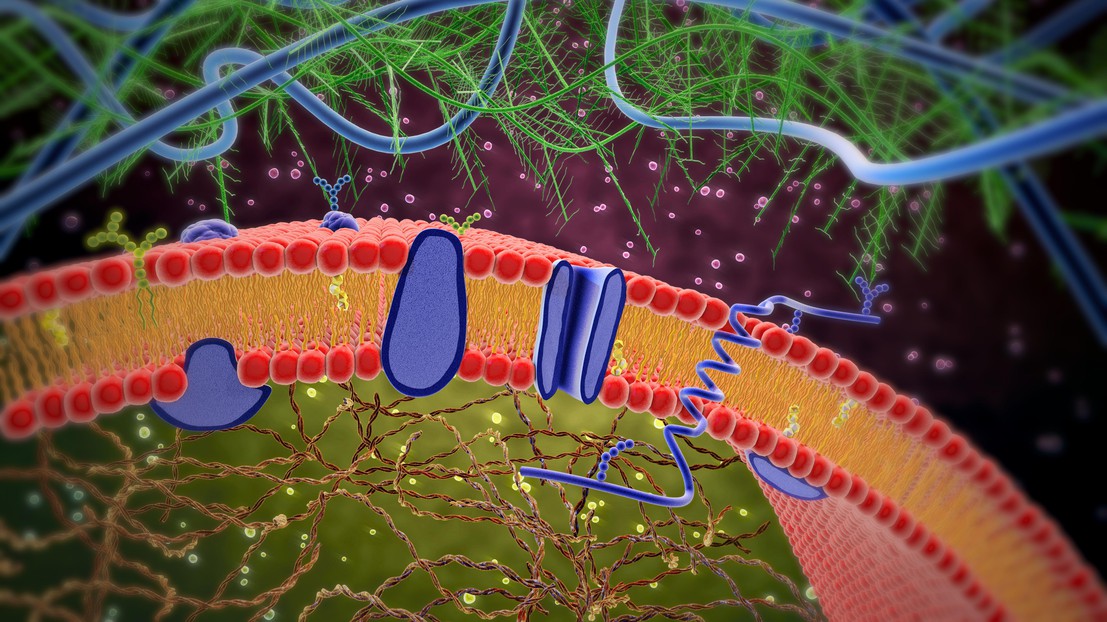
Cell membrane with membrane-bound and transmembrane receptors (Credit: Lars Neumann/iStock photos)
EPFL and US scientists have developed a computational method that can design synthetic cell receptors that can be used to isolate how drugs work in a cell, minimize or even altogether prevent side effects and redirect their action.
One of the challenges of modern pharmacology is specificity: despite their therapeutic effects, drugs can often have side effects. The biological basis of this has to do with the proteins and receptors that the drug targets and binds to. Many target receptors are connected to more than one biochemical pathway or – more commonly – the drug is not specific enough to exclusively bind one particular receptor.
A recent solution to this is to develop artificial target receptors that are only activated by ligands not found anywhere in the cell. The idea is that when “installed” in a cell, these artificial receptor-ligand pairs will only activate one biochemical pathway – sometimes a completely new one – without interfering with the cell’s other functions.
The field has mainly focused on cell receptors, and especially those located on the cell membrane. These receptors have enormous biomedical and pharmacological potential, as they translate extracellular signals into specific intracellular functions. But because they often bind to multiple intracellular proteins with interactions that are not currently very well understood, it has proven difficult to design synthetic receptors with novel signaling functions.
Now, scientists in Patrick Barth’s lab at EPFL, the Ludwig Institute for Cancer Research Lausanne and Baylor College of Medicine in the US have developed a powerful computational method to accurately model and engineer synthetic orthogonal receptor-ligand pairs that bind together and transmit biochemical signals in the cell with high selectivity.

The method integrates various aspects of synthetic computational biology (e.g. membrane protein homology modeling, ligand and protein docking) with techniques that can design membrane receptor–transmitter protein coupling even if there is no information on the structure of the various components or their interactions.
The scientists tested their method on the D2 dopamine receptor, which is heavily involved in various functions of the central nervous system, such as sensing, behavior, and movement. In addition, the D2 receptor is also connected to diseases such as Parkinson’s and Alzheimer’s, meaning that synthetic dopamine receptors with fine-tuned signaling properties would be powerful tools to better study biochemical signaling pathways and even develop gene therapies against neurodegenerative diseases in the future.
The synthetic receptor and its ligand bound together with high efficiency, and proved able to trigger the intended functions in the cell without interfering with its other, natural activities.
Finally, the designed receptor displayed distinct motifs in its amino acid sequence, which expands the “alphabet” for recognizing receptors and their ligands.
“This design approach can be used to reprogram cellular functions in cell-engineering applications,” says Patrick Barth. “For example, one could envision designing synthetic biosensors that redirect immune-inhibitory signals from tumor microenvironments into proliferative and activating cellular functions. Harnessing engineered immune cells – like chimeric antigen receptor T cells – with such biosensors is likely to improve their anti-tumor functions and lead to cancer control.”
Other contributors
- University of Lausanne
- Ludwig Institute for Cancer Research Lausanne Branch
- Baylor College of Medicine
- University of Montreal
National Institutes of Health (NIH)
NIH National Institute of General Medical Sciences
Leukemia and Lymphoma Society Translational Research Program
Cancer Prevention and Research Institute of Texas (Individual Investigator Research Award)
EPFL
Ludwig Institute for Cancer Research
M. Young, T. Dahoun, B. Sokrat, C. Arber, K. M. Chen, M. Bouvier, P. Barth. Computational design of orthogonal membrane receptor-effector switches for rewiring signaling pathways. PNAS 18 June 2018. DOI: 10.1073/pnas.1718489115