New study into leukemia offers clearer understanding of its biology
Blast cells in leukemia (iStock Photos)
EPFL scientists have made an extensive study in the cause of leukemia that could greatly improve the way we treat the disease.
Leukemia affects 350,000 people worldwide. It is a cancer of the white blood cells, which are the cells of the immune system and are produced by stem cells in the bone marrow. There are two types of chronic and two types of acute leukemia. One chronic type, chronic myeloid leukemia accounts for ~20% of all cases, and is caused by a mutated enzyme with unregulated activity. This enzyme comes in two sizes, the second of which is associated with acute lymphocytic leukemia. Being the focus of current leukemia treatment, understanding the role of the enzyme is critical. EPFL scientists have now used proteomics to compare the two forms of the enzyme extensively, uncovering a much clearer view of how it may give different forms of leukemia. The work, published in Leukemia, opens up possibilities for improved drug development and therapy.
Chronic myeloid leukemia is caused when two chromosomes in bone marrow stem cells swap specific parts. The resulting chromosome is called “Philadelphia”, and now contains a fused gene that produces an abnormal enzyme called Bcr-Abl.
This enzyme, which is constantly on, comes into two different sizes, or isoforms: the bigger (p210) is associated with chronic myeloid leukemia, while the shorter one (p190) is linked with acute lymphoblastic leukemia, a fast-developing form that is very difficult to treat leading to death of patients within a few months. The data on the differences of the two Bcr-Abl isoforms is inconsistent and confusing, meaning that we need more clarity as to its role in the development of this type of cancer.
The lab of Oliver Hantschel at EPFL, in close collaboration with the Proteomics Core Facility at the School of Life Sciences, used a proteomics approach to study the two isoforms of Bcr-Abl. Proteomics is a cutting-edge field that uses mass spectrometry analysis, which enables to map the entire set of proteins of an organism, termed the proteome.
The researchers looked at two factors: First, all of the ways each Bcr-Abl isoform interacts with other molecules in the cell, or its “interactome”. This allowed them to map out how the two isoforms work with other molecules.
Second, the team looked at the isoforms’ phosphoproteomics. This refers to a modification — phosphorylation— that many proteins, including Bcr-Abl undergo in the cell, whereby they are “turned on” with the addition of a phosphate group. Using mass spectrometry, the researchers were able identify and compare hundreds of phosphorylated proteins in leukemia cells.
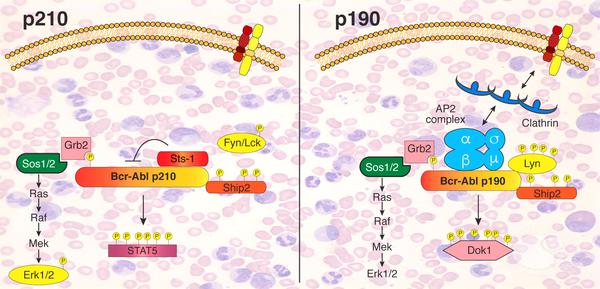
Scheme depicting the different signaling networks of the two Bcr-Abl isoforms determined in this study (credit: Sina Maren Reckel/EPFL)
By comparing the two, the researchers were able to map out that there are “surprisingly large” differences between the interactome and phosphoproteome of the two isoforms, even though they are both “turned on” in much the same way. These differences in interactions and activation could be related to how each isoform drives an entirely different form of leukemia, and opens up possibilities for targeting Bcr-Abl in more efficient and effective ways to treat the disease.
This study involved a collaboration of EPFL’s Swiss Institute for Experimental Cancer Research (ISREC), its Proteomics Core Facility. It was funded by the ISREC Foundation, the Swiss National Science Foundation (SNSF) and the National Center of Competence in Research (NCCR) in Chemical Biology.
Reference
Sina Reckel, Romain Hamelin, Sandrine Georgeon, Florence Armand, Qinfang Jolliet, Diego Chiappe, Marc Moniatte, Oliver Hantschel. Differential signaling networks of Bcr-Abl p210 and p190 kinases in leukemia cells defined by functional proteomics.Leukemia 23 January 2017. DOI: 10.1038/leu.2017.36