“I was lucky to have found this project, along with the right people”
© Alain Herzog / 2021 EPFL
Master’s student in computer science, Quentin Juppet has just published a paper on life sciences as the study’s lead author. With a cross-disciplinary team of biologists, engineers and computer scientists, they develop an algorithm for distinguishing mouse cells from human ones in a tissue sample. Their work will be useful in the field of cancer research.
Quentin Juppet, originally from France’s Savoy region, became interested in computer science at an early age. He was just 15 when he took his first programing classes, given by EPFL professors. “I started by taking a MOOC on the C++ programming language, and the next year I took one on Java. These online courses were how I learned about EPFL. Then when I came here for my Bachelor’s degree, I got to meet the professors in person and even ended up becoming an assistant for one of them,” he says.
An innovative image-processing tool
Juppet is now in the second year of his Master’s degree at EPFL’s School of Computer and Communication Sciences and has just published his first study, carried out as part of a cross-disciplinary team of biologists, engineers and computer scientists from across EPFL.
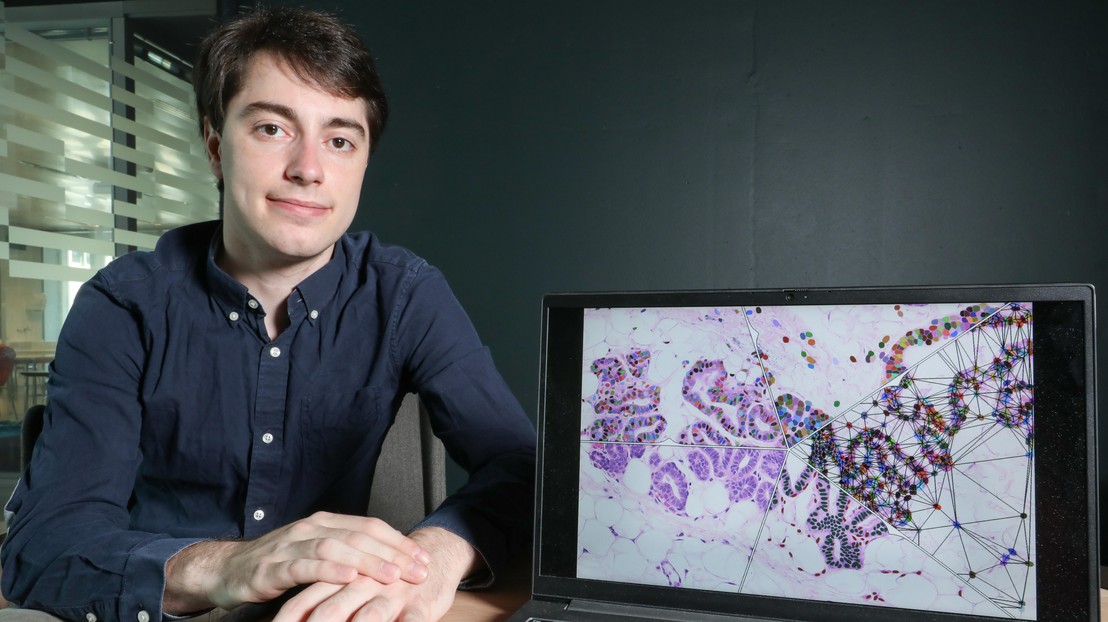
The team developed a machine-learning algorithm that can distinguish mouse cells from human ones in a tissue sample, which is particularly useful for cancer research. Oncologists typically study tumors by grafting human cells onto mice, but then the problem is telling the two kinds of cells apart. That usually involves several rounds of fluorescence staining and analyzing many tissue samples before finding the human cells. However, Juppet’s program simplifies all that by automating the cell-classification process.
To develop the program, the researchers first came up with a list of features that characterize each type of cell. “We worked with Fabio De Martino, a PhD student at Cathrin Brisken’s lab in the School of Life Sciences, to translate biological data like size and shape into numerical values. In addition to cell-type information, we also factored in the cells’ surroundings by including parameters on contiguous cells,” says Juppet. Based on these data, the machine-learning algorithm can spot which cells in a sample come from humans and which ones come from mice. What’s more, it can be configured to identify other types of cells. The program, called Single Cell Classifier (SCC), is available in open-source format as a plug-in for the ImageJ image-processing software.

Scaling up a class project
At first, Juppet planned to carry out just a four-week class project on the program. “The project was for a machine-learning class I took in the first year of my Master’s degree. We worked in groups, and this topic was suggested by Prof. Brisken’s lab – but we didn’t have enough time to finish something that would be genuinely useful for research,” says Juppet. He therefore decided to turn the class project into his semester project and worked on it for a few more months.
“My supervisor for the semester project was Dr. Daniel Sage, a senior scientist at EPFL’s Biomedical Imaging Laboratory in the School of Engineering,” says Juppet. “He advised me on algorithms and on imaging in general.” Juppet carried out the project through EPFL’s newly opened Center for Imaging (ECI), working with Dr. Martin Weigert, a group leader at EPFL’s School of Life Sciences, and Olivier Burri, an image analyst at the BIOP bioimaging platform where the images used in Juppet’s study were captured. The research team was able to complete the project in record time, eventually leading to the biology journal publication. “I was lucky to have gotten involved in this project and to have found the right people to work with,” says Juppet. “I also had access to professors who offered their guidance and to EPFL’s research ecosystem, where everything I needed was available.”
See also:New Imaging Center Pools the Know-How of Five EPFL Schools
A taste for research
Juppet took the research even further for his Master’s project, working with a local business to fine-tune his image-processing algorithm. “I completed my Master’s project at Lunaphore, an EPFL spin-off where I worked part-time during my studies – it was important for me to do something hands-on alongside my classwork,” he says. Juppet will soon graduate and is planning to do a PhD. “First I want to work for Lunaphore full-time for at least a year. Then, who knows, I might just stay in academia. I have to admit that this first journal publication has given me a taste for research,” he says.
Juppet, Q., De Martino, F., Marcandalli, E., Weigert, M., Burri, O., Unser, M., Brisken, C., and Sage, D. “Deep Learning Enables Individual Xenograft Cell Classification in Histological Images by Analysis of Contextual Features.” J Mammary Gland Biol Neoplasia (2021). https://doi.org/10.1007/s10911-021-09485-4

