Genetic observation reveals a bone-weakening mechanism
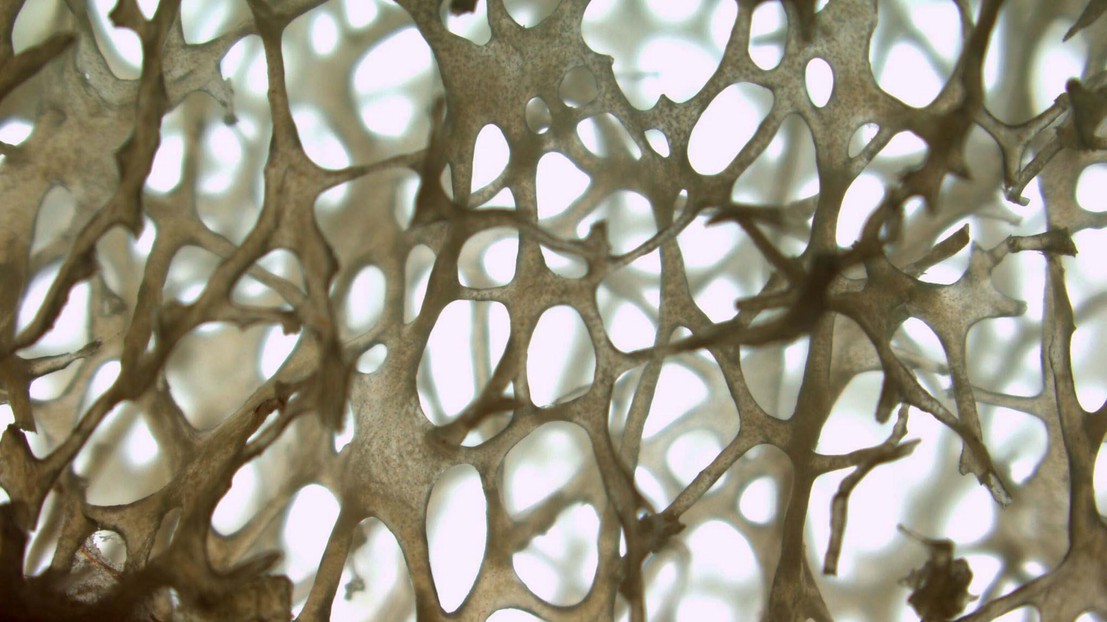
Bone structure and firmness partly rely on some genes. © Inserm - Daniel Chappard
An EPFL research team has used a novel method to identify a gene involved in bone building. Their results appear today in the advance online edition of the scientific journal Cell.
“Real life genetics” works. This research method involves observing physiological traits or metabolic disease in a large population of “wild-type” mice (those which have not been genetically modified), and then isolating the genes that could be responsible. Because it’s complex and expensive, the method is rarely used, but it has nonetheless enabled EPFL researchers to obtain interesting results.
An article published today* in the online edition of the journal Cell describes how the method was used to reveal a dysfunctional bone mineralization process. “We compared the genotype of mice with a fragile bone structure with mice that were more robust,” explains Evan Williams, a scientist in EPFL’s Laboratory for Integrative and Systemic Physiology (LISP). “By observing their genes, we were able to identify the one responsible for alkaline phosphatase (ALPL), whose malfunction leads to various forms of bone disease.”
A factor in osteoporosis
Scientists were already familiar with hypophosphatase, a human disease caused by the absence of this gene that leads to extreme skeletal fragility from a very young age. “With this new discovery we can conclude that a defect in the gene expression for alkaline phosphatase affects bone strength in mature adults. In fact, fractures are the primary cause for hospitalization in the elderly,” says LISP director Johan Auwerx. Pénélope Andreux, a co-author of the study, adds that the gene has other jobs in the cell as well: “it plays an important role in vitamin B6 absorption, which is involved in protein metabolism,” she explains.
Very different characteristics
These results suggest that new therapeutic avenues might be possible for preventing osteoporosis, by acting on the enzymes associated with the expression of this gene. But that’s not all. The study of this population of mice wasn’t limited to their bones: no less than 140 parameters, such as body weight, running speed and fur color, were measured, and the results were often astonishing.
For example, in a single population and without any added stimulation, some mice spontaneously ran two km every night, while others ran twelve. And some mice had up to 6.7 times better glucose tolerance than others.
What could explain these differences? “Our results, which we have made available to the scientific community, will allow specialists in various fields to make associations between the genome of these mice and their physical and behavioral characteristics,” says Auwerx. “Then it will be a matter of establishing correlations with the expression of the same genes in humans.” Observing these mice populations will thus lead to many discoveries.
* Pénélope A. Andreux, Evan G. Williams et al., Systems Genetics of Metabolism : The Use of the BXD Murine Reference Panel for Multiscalar Integration of Traits. Cell, http://dx.doi.org/10.1016/j.cell.2012.08.012